Figures & data
Figure 1 RNAi/miRNA pathway schematization and major challenges for naked siRNA delivery in vivo. (A) Schematization of RNA interference (RNAi): non-translated double-stranded RNA (dsRNA) molecules called small interfering RNA (siRNA), of exogenous or endogenous origin, post-transcriptional regulate gene-expression through a sequence specific degradation of target messenger RNA (mRNA). [1] Longer siRNA molecules (dark green) are cleaved by the nuclease Dicer and [2] incorporated into a multiprotein RNA-inducing silencing complex (RISC). [2] The duplex RNA is unwound leaving the anti-sense strand (light green). [3] to guide RISC to complementary mRNA (red) for subsequent endonucleolytic cleavage and gene silencing. [4] Short hairpin RNAs (shRNA) (violet) are sequences of RNA encoded by specific genes; they are introduced into the nucleus, transcribed and transported into the cytoplasm where they follow the same fate of siRNA. (B) miRNA processing: microRNA (blue) are considered as the “endogenous substrate” of the RNAi machinery. They are trascribed by RNA-Pol III in long primary transcripts (pri-miR), then processed within the nucleus into precursor miRNA (pre-miRNA) by the RNase III enzyme Drosha-DGCR8. Pre-miRNA hairpins are exported from the nucleus in a process involving the nucleo-cytoplasmic shuttle Exportin-5 (Exp.5). In cytoplasm, the pre-miRNA hairpin is cleaved by Dicer and loaded into RISC as for siRNA. miRNAs often share only partial complementarity with target mRNAs, usually in the 3′UTR, acting mainly as a translational repressors. (C) “Naked” siRNA pitfalls. In the box are reported the major obstacles for therapeutic efficacy of siRNA without modifications (“naked”). See text for details.
![Figure 1 RNAi/miRNA pathway schematization and major challenges for naked siRNA delivery in vivo. (A) Schematization of RNA interference (RNAi): non-translated double-stranded RNA (dsRNA) molecules called small interfering RNA (siRNA), of exogenous or endogenous origin, post-transcriptional regulate gene-expression through a sequence specific degradation of target messenger RNA (mRNA). [1] Longer siRNA molecules (dark green) are cleaved by the nuclease Dicer and [2] incorporated into a multiprotein RNA-inducing silencing complex (RISC). [2] The duplex RNA is unwound leaving the anti-sense strand (light green). [3] to guide RISC to complementary mRNA (red) for subsequent endonucleolytic cleavage and gene silencing. [4] Short hairpin RNAs (shRNA) (violet) are sequences of RNA encoded by specific genes; they are introduced into the nucleus, transcribed and transported into the cytoplasm where they follow the same fate of siRNA. (B) miRNA processing: microRNA (blue) are considered as the “endogenous substrate” of the RNAi machinery. They are trascribed by RNA-Pol III in long primary transcripts (pri-miR), then processed within the nucleus into precursor miRNA (pre-miRNA) by the RNase III enzyme Drosha-DGCR8. Pre-miRNA hairpins are exported from the nucleus in a process involving the nucleo-cytoplasmic shuttle Exportin-5 (Exp.5). In cytoplasm, the pre-miRNA hairpin is cleaved by Dicer and loaded into RISC as for siRNA. miRNAs often share only partial complementarity with target mRNAs, usually in the 3′UTR, acting mainly as a translational repressors. (C) “Naked” siRNA pitfalls. In the box are reported the major obstacles for therapeutic efficacy of siRNA without modifications (“naked”). See text for details.](/cms/asset/9b1772a2-08da-4fc5-af9d-ad5016fd6fde/dijn_a_23696_f0001_c.jpg)
Table 1 Problems of Naked siRNA for clinical applications
Figure 2 Encapsulation technologies for siRNA delivery in vivo: nanoparticles strategies and advantages.
Readapted from Shim et al.Citation165
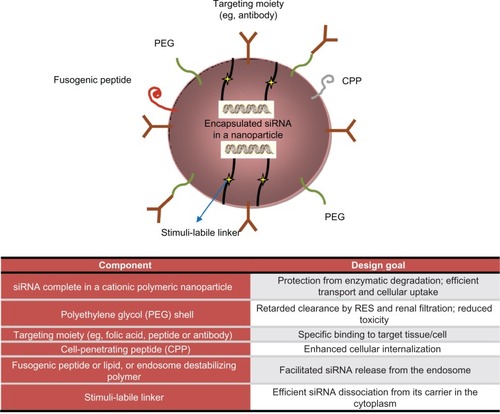
Table 2 Delivery strategies for siRNA. Advantages and pitfalls
Table 3 Nanoparticles in siRNA-based delivery
Figure 3 Obstacles of Nanoparticles-based siRNA delivery in vivo. After administration into blood circulation the siRNA-nanoparticles (A) must avoid rapid degradation by plasma components (eg, cellular and humoral arm of the immune system) and sequester by negatively charged serum protein. (B) Then they need to escape renal filtration and/or clearance by the reticuloendothelial system (RES). (C) To reach the target cells they must overcome the capillary endothelium through an extravasation process and (D) overcome the extracellular matrix (ECM): a dense network of polysaccharides and fibrous proteins, rich in macrophages, which can obstacle the transport of nanoparticles. (E) Furthermore these particles must be taken up into the cells, usually bound to cellular receptors and transported into the cytoplasm through a receptor mediated endocytosis process. (F) Inside the cells the particles need to escape the endosome; (G) thus unpackage and release the siRNA to the RNA interference (RNAi) machinery.
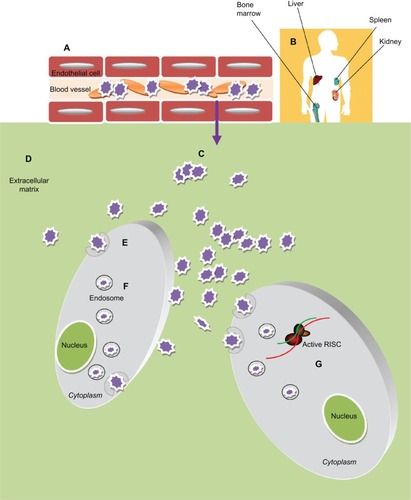
Table 4 Ongoing clinical trials in cancer and other diseases