Figures & data
Table 1 Cell/Donor Information
Figure 1 PLGA/pDNA-NPs were taken up by astrocytes but failed to facilitate exogenous gene expression. Luciferase reporter pDNA encapsulated in PLGA-NPs was used to assess PLGA-mediated gene delivery (A). HEKs or primary human astrocytes were treated with 120 µg PLGA/pDNA-NPs for 2, 3, and 6 days. Luciferase reporter assay was used to verify exogenous gene expression. Data are graphed as mean ± SD from a representative experiment with duplicated measurements from triplicate wells. Two additional experiments illustrated the same trend. (B) Primary human astrocytes were treated with YOYO-pDNA encapsulated in PLGA-NPs (60 or 120 µg) and imaged at 6, 24, 48 and 96 hours using fluorescent microscopy to gauge PLGA-NP-mediated pDNA delivery to the cytoplasm.
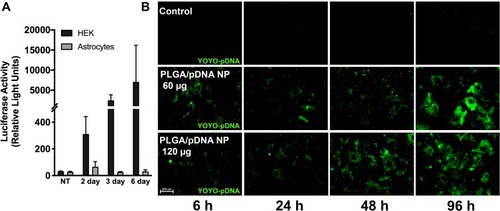
Figure 2 Co-transfection of PLGA/pDNA-NPs with A5P10 established an efficient and biocompatible mechanism for astrocyte gene therapy. Primary human astrocytes (A and B) or neurons (C) were treated with luciferase pDNA encapsulated in PLGA-NPs (60 or 120 µg) alone or with A5P10 polymer (1 or 2 µg) for 48 hours. (A) Luciferase activity was quantified using luciferase expression assay. The x-axis was broken to indicate the scale in luciferase activity, ie, bottom (0–1 RLU x 104) and the top (10–200 RLU x 104). Statistical significance was determined via two-way ANOVA and Tukey’s post hoc test. (B and C) Cytotoxicity was measured by LDH assay and normalized to control for each donor. Data represent mean ± SEM in 3–4 donors. Statistical significance was determined via one-way ANOVA followed by Tukey’s post hoc (**p<0.01, ***p<0.001).
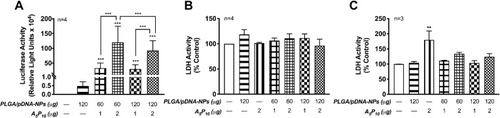
Table 2 NP Characterization
Figure 3 A5P10 increased PLGA/pDNA-NP-mediated uptake, intracellular trafficking, and nuclear delivery of pDNA. Primary human astrocytes were treated with PLGA/pDNA-NPs (60 or 120 µg) alone or in combination with 2 µg A5P10 for 24 hours. A5P10 coupled with YOYO-pDNA was used as a positive control. Confocal images (A) and colocalization analysis (B) were achieved using YOYO-pDNA and Hoechst nuclear dye. White arrows indicate YOYO and Hoechst colocalization. Scatter regions 1 and 2 depict pixels for YOYO and Hoechst channels, respectively, and scatter region 3 determines the pixels colocalizing between the two channels. (C) Graphs report mean ± SD for calculated total YOYO pixels divided by total pixels per image and (D) weighted colocalization coefficients in relation to total Hoechst pixels per image from a minimum of 8–10 images of triplicate wells per condition. Statistical significance was determined via one-way ANOVA followed by Tukey’s post hoc test (**p<0.01, ***p<0.001).
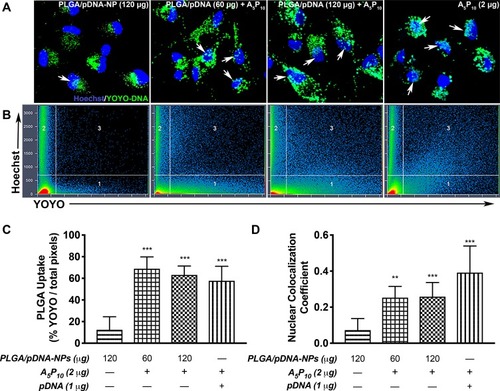
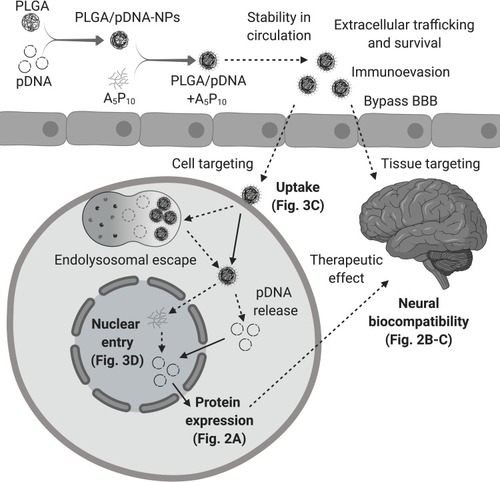
Figure 4 Obstacles for gene delivery to CNS. A successful gene delivery system to the CNS must achieve optimal trafficking and survival in the periphery including stability in circulation and evasion of the immune system. A critical dilemma for these constructs is bypassing the BBB in order to reach the tissue or cell of interest. Ensuring specificity is also of heightened importance for gene therapy applications. Once targeting is accomplished, delivery systems must overcome a variety of obstacles for intracellular trafficking including uptake, endolysosomal escape, nuclear entry, DNA release, and finally, expression of the therapeutic target. Moreover, systemic biocompatibility is an essential objective. Our report demonstrates (solid lines) PLGA/pDNA+A5P10 was able to facilitate enhanced uptake and intracellular trafficking (), nuclear entry of pDNA (), and exogenous gene expression () while maintaining biocompatibility in astrocytes and neurons (). To translate this system for successful gene therapy, additional investigations are warranted to determine the potential of PLGA/pDNA+A5P10 to overcome the remaining barriers and to further elucidate the intracellular mechanisms mediating PLGA/pDNA+A5P10 optimization (dashed lines). (Created with BioRender.com).