Figures & data
Table 1 Representative Conformational Changes in Proteins Following Complexation with NPs
Table 2 Structural Changes in Model Proteins on Various Surfaces of Carbon Nanotubes
Table 3 Secondary Structural Changes Depending on the Surface Characteristics
Figure 1 Size comparison between proteins and NPs.
Notes: The sizes of NPs are compared to those of various proteins. The codes in brackets are those from the PDB database. Adapted with permission from Kopp M, Kollenda S, Epple M. Nanoparticle-Protein Interactions: Therapeutic Approaches and Supramolecular Chemistry. Acc Chem Res. 2017; 50(6):1383–1390. Copyright (2019) American Chemical Society.Citation50
Abbreviations: HSA, human serum albumin; IgG, immunoglobulin G.
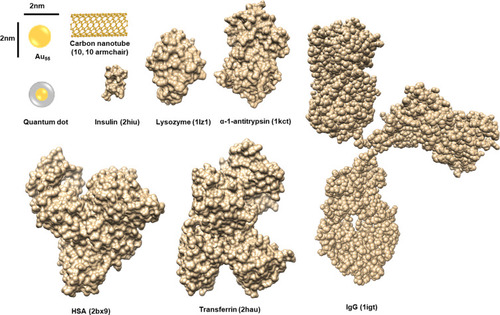
Figure 2 Extracellular and intracellular events caused by the structural changes of corona proteins.
Notes: The interaction of NPs with proteins can induce variety of signal modulations and toxic effects in biofluids and in cells. Various physicochemical properties of NP systems basically contribute to the corona formation and structural changes of proteins. The conformational change is a dynamic process and the composition of corona proteins on NPs can be changed according to the surrounding environment. The reversible or irreversible changes of protein structures can perturb the downstream signaling, which may consequently be harmful to the host. The characteristics of corona formation may be expected to be different between extracellular and intracellular spaces. The internalized NPs by various ways such as receptor mediated internalization and endocytosis by charge can produce many toxic situations directly through their own chemical characteristics and/or indirectly through corona formation.
Abbreviations: ER, endoplasmic reticulum; ROS, reactive oxygen species.
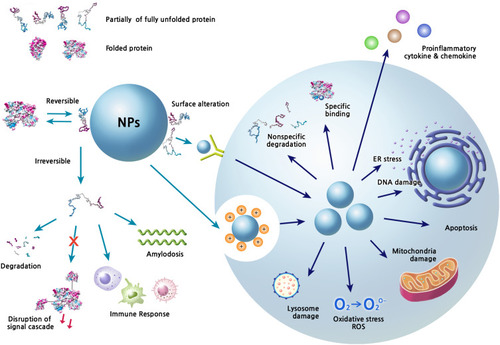
Figure 3 The preference of secondary structural elements for MWCNT binding.
Notes: About 750 cellular proteins were monitored for binding to MWCNT by mass spectrometry-based proteomics. The secondary structures of the bound proteins were analyzed using the 3D structures deposited in the Protein Data Bank. 269 proteins out of identified 778 proteins were used for the analysis. The proportion of α-helix and β-sheet for each protein was quantified. The protein classes among the CNT-binding proteins are represented with bubbles. The most abundant protein classes contained cytoskeletal proteins, endosomal proteins, and heat shock proteins. The bubble size represents the relative binding affinity to MWCNTs. Adapted from Nanomedicine: Nanotechnology, Biology and Medicine, 9(5), Cai X, Ramalingam R, Wong HS, Cheng J, Ajuh P, Cheng SH, Lam YW. Characterization of carbon nanotube protein corona by using quantitative proteomics. Nanomedicine. 583–593, Copyright (2013), with permission from Elsevier.Citation87
Abbreviation: MWCNT, multi-walled carbon nanotube.
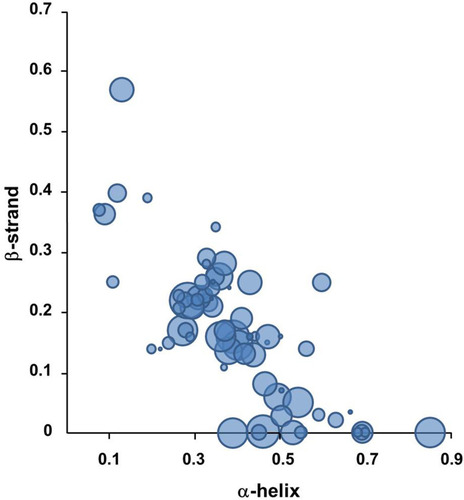
Figure 4 Secondary structures of protein-NP complexes (unpublished data).
Notes: PBS represents the reference CD curve without NPs, measured only in phosphate-buffered saline (pH 7.4). All other curves were measured with NPs in phosphate-buffered saline (pH 7.4). (A) Human serum albumin, (B) α-1-antitrypsin, (C) transferrin, (D) lysozyme, (E) fibrinogen (α, β, r).
Abbreviations: CD, circular dichroism; NP, nanoparticle.
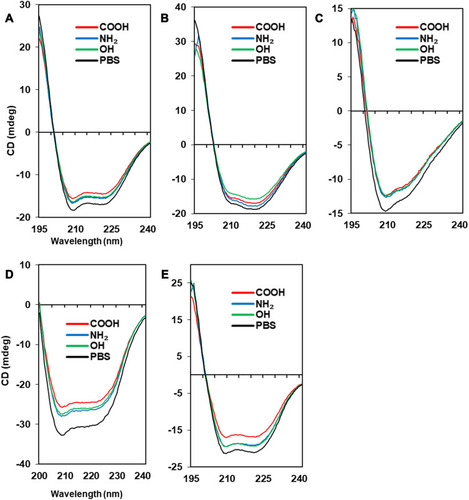
Figure 5 The Tm curves of protein-NP complexes.
Notes: PBS represents the reference Tm curve without NPs. (A) Albumin (PDB code: 2bx8), (B) α-1-antitrypsin (PDB code: 1kct), (C) transferrin (PDB code: 2hau), (D) lysozyme (PDB code: 1lz1), (E) fibrinogen (α, β, r, PDB code: 3ghg). The structure of each protein was obtained from PDB database (http://www.rcsb.org). The Coulumb parameters were ε = 4r and thresholds ± 5.93 kcal/mol⋅e. Negative charge is indicated in red, positive in blue, and neutral in white. Surface charge was calculated using UCSF Chimera.
Abbreviations: CD, circular dichroism; NP, nanoparticle; Tm, melting temperature.
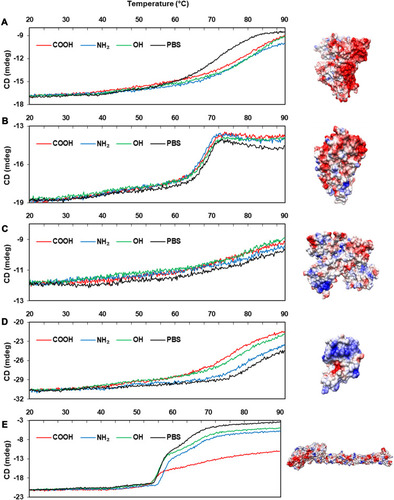
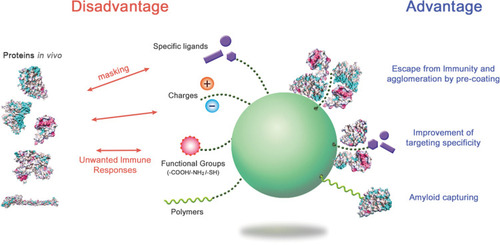
Figure 6 The advantages and disadvantages of protein corona formation.
Notes: By regulating the protein coronas through appropriate surface modification and NP selection, the biological behavior of NPs in the body could be improved. Structural studies on the corona proteins in NP systems may provide insight into how to handle the NPs.
Abbreviation: NPs, nanoparticles.