Figures & data
Figure 1 Transmission electron microscopic images of selenium nanoparticles obtained at different Spirulina polysaccharide concentrations at the reaction time of 24 hours: (A) 0 mg/L; (B) 1 mg/L; (C) 5 mg/L; (D) 10 mg/L; (E) 50 mg/L; (F) 100 mg/L.
Note: Scale bar 500 nm.
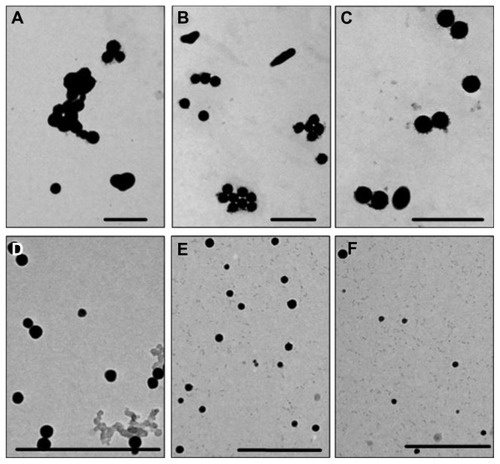
Figure 2 (A) X-ray diffraction pattern, (B) Raman spectrum, and (C) scanning electron microscopy and energy dispersive X-ray analysis of selenium nanoparticles functionalized by Spirulina polysaccharide.
Abbreviations: au, absorbance units; C, carbon; O, oxygen; Se, selenium.
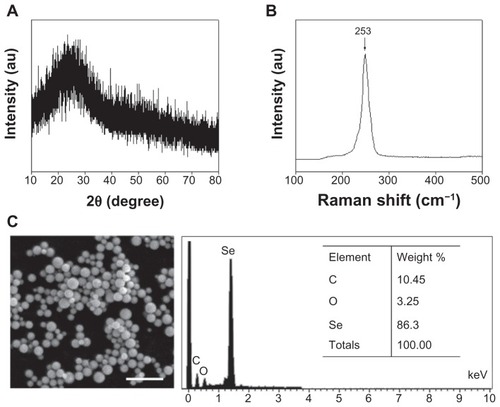
Figure 3 Fourier transform infrared spectra of (A) Spirulina polysaccharide and (B) selenium nanoparticles functionalized by Spirulina polysaccharide.
Abbreviation: au, absorbance units.
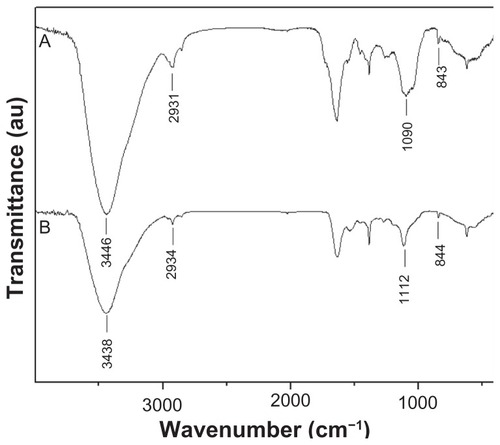
Figure 4 (A) Size distribution of selenium nanoparticles obtained at different Spirulina polysaccharide concentrations at the reaction time of 24 hours and (B) time-course of size distribution of selenium nanoparticles functionalized by Spirulina polysaccharide obtained at 50 mg/L of Spirulina polysaccharide.
Note: Each value represents mean ± standard deviation (n = 3).
Abbreviations: d, days; SPS, Spirulina polysaccharide.
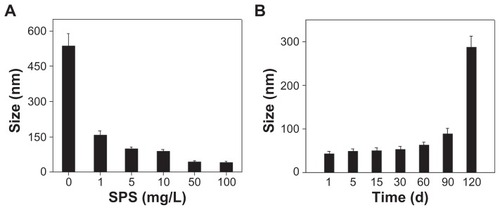
Figure 5 Functionalization of selenium nanoparticles with Spirulina polysaccharide.
Abbreviations: Se, selenium; SeO32−, selenite; SeNPs, selenium nanoparticles; SPS, Spirulina polysaccharide.
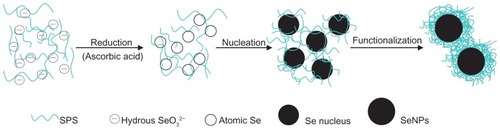
Figure 6 Cellular uptake of selenium nanoparticles functionalized by Spirulina polysaccharide by A375 cells. Cells were exposed to selenium nanoparticles functionalized by Spirulina polysaccharide or selenium nanoparticles at different concentrations for 72 hours. Cells without treatment were used as control. Intracellular selenium concentrations were determined with inductively coupled plasma atomic emission spectroscopy method.
Note: Each value represents mean ± standard deviation (n = 3).
Abbreviations: Se, selenium; SeNPs, selenium nanoparticles; SPS-SeNPs, selenium nanoparticles functionalized by Spirulina polysaccharide.
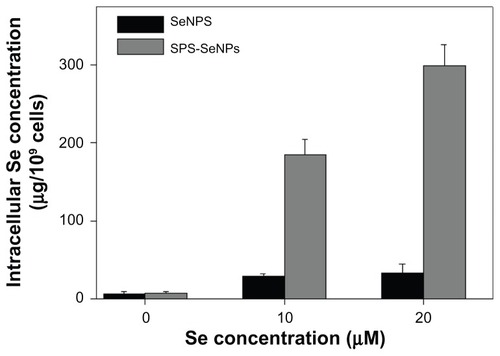
Figure 7 (A) Half maximal inhibitory concentration value of various cancer and normal cell lines after incubation for 72 hours with selenium nanoparticles functionalized by Spirulina polysaccharide and (B) morphological changes in A375 cells observed by phase contrast microscopy (magnification 100×).
Notes: Each value represents mean ± standard deviation (n = 3). IC50 values were based on selenium concentration as determined by inductively coupled plasma atomic emission spectroscopy method.
Abbreviations: IC50, half maximal (50%) inhibitory concentration; SPS-SeNPs, selenium nanoparticles functionalized by Spirulina polysaccharide.
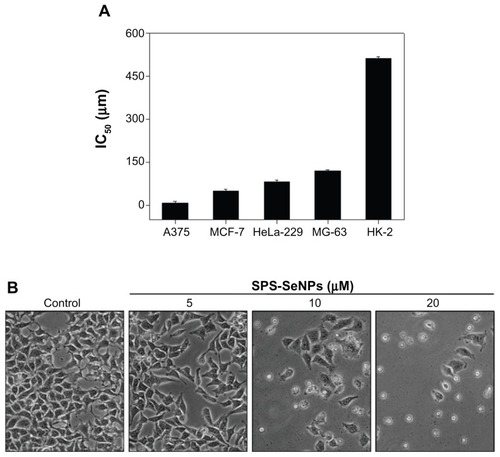
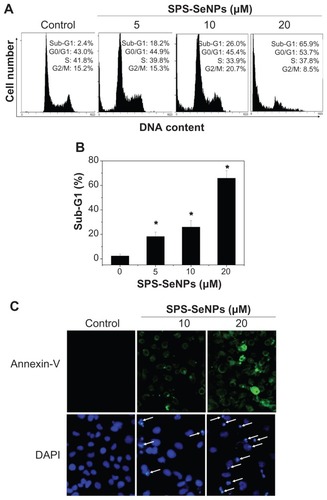
Figure 8 Induction of apoptosis by selenium nanoparticles functionalized by Spirulina polysaccharide. (A) Flow cytometric analysis of cell cycle distribution of A375 cells treated for 24 hours. (B) Quantitative analysis of selenium nanoparticles functionalized by Spirulina polysaccharide induced apoptotic cell death by measuring the sub-G1 cell population. Significant difference between selenium nanoparticles functionalized by Spirulina polysaccharide treatment and control is indicated at P < 0.05 level. (C) Representative images of phosphatidylserine translocation (upper panel) and nuclear condensation (lower panel) in A375 cells exposed to selenium nanoparticles functionalized by Spirulina polysaccharide for 24 hours as examined by Annexin-V-FLUOS labeling assay (magnification 600×) and 4′,6-diamidino-2-phenylindole staining assay (magnification 600×), respectively.
Note: *P < 0.05.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DNA, deoxyribonucleic acid; SPS-SeNPs, selenium nanoparticles functionalized by Spirulina polysaccharide.