Figures & data
Figure 1 Three dimensional model of the P2 monomer from Hib (panel A) showing the target of our study, loop L7; signal transduction pathways analyzed in this study and their inhibition with complementary peptides (panel B); Kyte-Doolittle hydropathy plots of L7 and complementary peptides (panel C).
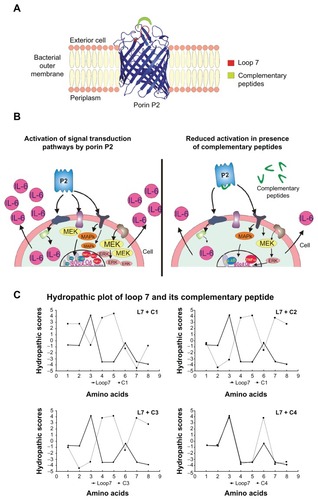
Table 1 Peptide sequences
Figure 2 MEK1-MEK-2/MAPK activation in U937 cells in response to Hib porin (13 nmol/ml) or peptides (130 nmol/mL). In some assays, the complementary peptides C1, C2, C3 and C4 were preincubated for 60 minutes at 37°C with Hib porin or loop L7 and then used for cell stimulation. U937 cells (3 × 106 cells/mL) were stimulated for 10 min, lysed and immunoprecipitated with anti-phospho-specific form antibodies of each enzyme. The immunoprecipitates were subjected to SDS-PAGE, blotted onto PVDF membrane and reacted with specific HRP conjugated antibodies.
Notes: Gels were scanned for densitometry analysis with the Sigma Gel Software, and the ratio of the value for each stimulation to the value for protein P2, which was taken as 100%, is shown. The data are averages from three different experiments; the error bars indicate the standard errors of the means.
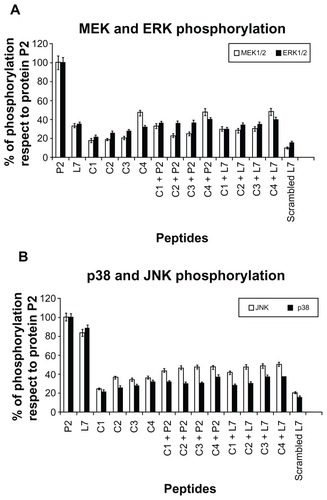
Figure 3 TNF-α (panel A) and IL-6 (panel B) release induced by loop L7 complementary peptides. U937 cells (3 × 106 cells/mL) were stimulated with Hib porin (13 nmol/mL) or peptides (130 nmol/mL) for 24 hours at 37°C in 5% CO2; in some assays, the complementary peptides C1, C2, C3 and C4 were preincubated for 60 minutes at 37°C with Hib porin or loop L7 and then used for U937-stimulation.
Notes: The results shown are the average of three independent experiments; the error bars indicate the standard errors of the means.
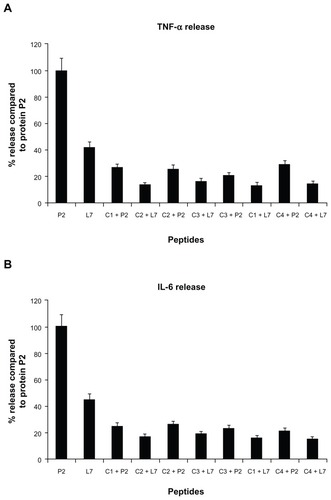
Figure 4 AP-1 activation in presence of loop L7 complementary peptides. U937 cells (10 × 106 cells/mL) were stimulated with Hib porin (13 nmoL/mL) or peptides (130 nmol/mL) for 1 hour at 37°C in 5% CO2; in some assays, the complementary peptides C1, C2, C3 and C4 were preincubated for 60 min at 37°C with Hib porin or loop L7 and then used for U937-stimulation.
Notes: Cell lysates (10 μg/mL) were tested for binding of the activated c-Fos or c-Jun subunits to an AP-1 consensus sequence using the Trans-Am AP-1 ELISA kit. The experiment was performed in the presence of soluble wild-type or mutated consensus oligonucleotides. The results are expressed as specific binding (absorbance measured in the presence of the mutated oligonucleotide minus that measured in the presence of the wild-type oligonucleotide). The results are shown as means ± standard errors of triplicate determinations.
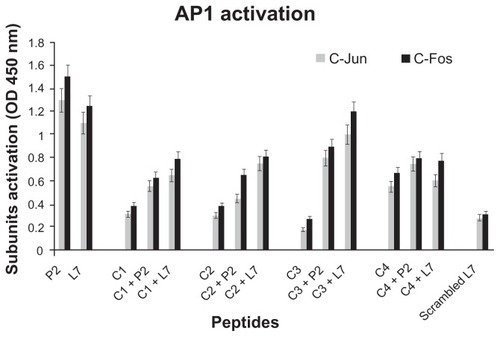
Figure 5 NF-κB activation in presence of loop L7 complementary peptides. U937 cells (10 × 106 cells/mL) were stimulated with Hib porin (13 nmol/mL) or peptides (130 nmol/mL) for 1 hour at 37°C in 5% CO2; in some assays, the complementary peptides C1, C2, C3 and C4 were preincubated for 60 minutes at 37°C with Hib porin or loop L7 and then used for U937-stimulation.
Notes: Cell lysates were tested for binding of the activated p50 or p65 subunits to an NF-κB consensus sequence using the Trans-Am NF-κB ELISA kit. The experiment was performed in the presence of soluble wild-type or mutated consensus oligonucleotides. The results are expressed as specific binding (absorbance measured in the presence of the mutated oligonucleotide minus that measured in the presence of the wild-type oligonucleotide). The results are shown as means ± standard errors of triplicate determinations.
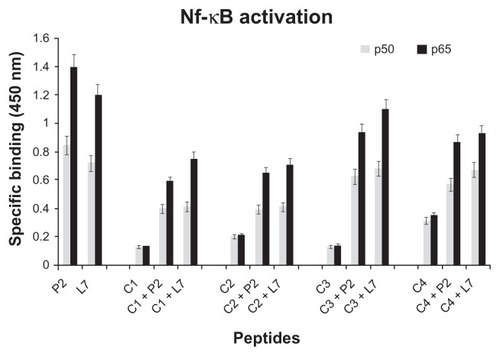
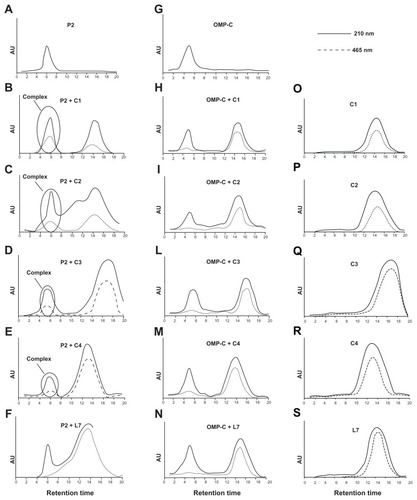
Figure 6 Binding of complementary peptides to porin P2 and OMPC by gel filtration. Peptides C1, C2, C3, C4 and L7 (panels O–S) were mixed to porin P2 (panels A–F) or porin OMPC (panels G–N) with a molar ratio protein/peptide of 1/50.
Notes: The binding mixture was held for 1 hour at 37°C and then was loaded on a PD-10 column containing Sephadex G-25 medium. The collected fractions were analyzed by UV spectrometry and the chromatographic profile obtained at 210 nm and 465 nm is shown.