Figures & data
Figure 1 Synthesis pathway of Y-shaped mPEG-P(LA-co-GA)2 copolymers.
Abbreviations: DMAP, 4-dimethylaminopyridine; Sn(Oct)2, stannous octoate; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide).
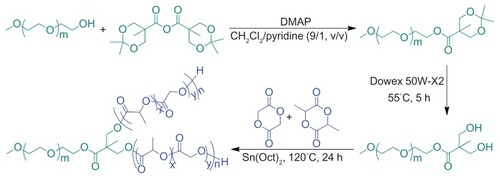
Figure 2 Schematic illustration of preparation of micellar/vesicular nanomedicines based on DOX and amphiphilic Y-shaped copolymers.
Abbreviations: DOX, doxorubicin; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide); PB, phosphate buffer.
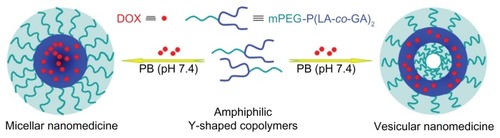
Table 1 Characterizations of mPEG-P-(LA-co-GA)2 copolymers
Figure 3 (A) 1H NMR and (B) FT-IR spectra of (a) mPEG113-(OH)2 and (b) mPEG113-b-P(LA12-co-GA9)2.
Abbreviations: 1H NMR, proton nuclear magnetic resonance; FT-IR, Fourier transform infrared; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide).
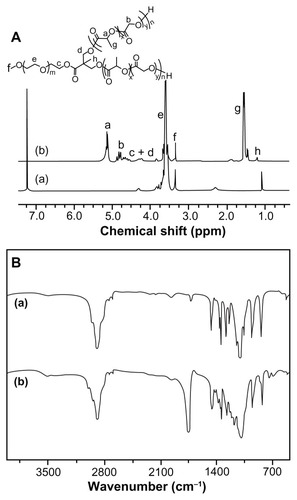
Figure 4 GPC chromatograms of (a) mPEG-(OH)2, (b) mPEG-P(LA4-co-GA9)2, (c) mPEG-P(LA12-co-GA9)2, (d) mPEG-P(LA24-co-GA8)2, and (e) mPEG-P(LA45-co-GA15)2.
Abbreviations: GPC, gel permeation chromatography; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide); Mn, number-average molecular weight; PDI, polydispersity index.
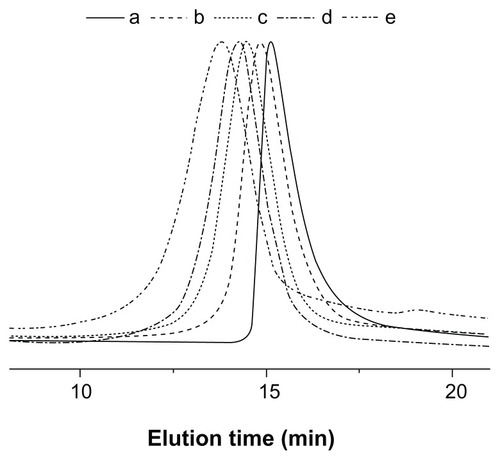
Table 2 Properties of mPEG-P(LA-co-GA)2 nanoparticles
Figure 5 (A) Excitation spectra of pyrene in aqueous solution of mPEG-P(LA12-co-GA9)2 at different concentrations (λem = 390 nm); (B) the intensity ratio (I336.5/I334) as a function of concentration of (a) mPEG-P(LA4-co-GA9)2, (b) mPEG-P(LA12-co-GA9)2, (c) mPEG-P(LA24-co-GA8)2, and (d) mPEG-P(LA45-co-GA15)2.
Abbreviations: mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide).
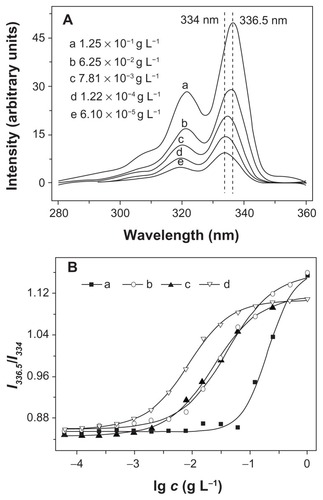
Figure 6 Typical TEM micrographs of (A) mPEG-P(LA12-co-GA9)2 and (C) mPEG-P (LA24-co-GA8)2, and Rh of (B) (a) mPEG-P(LA4-co-GA9)2, (b) mPEG-P(LA12-co-GA9)2, and (c) mPEG-P(LA45-co-GA15)2, and (D) mPEG-P(LA24-co-GA8)2.
Abbreviations: TEM, transmission electron microscopy; Rh, hydrodynamic radius; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co- glycolide).
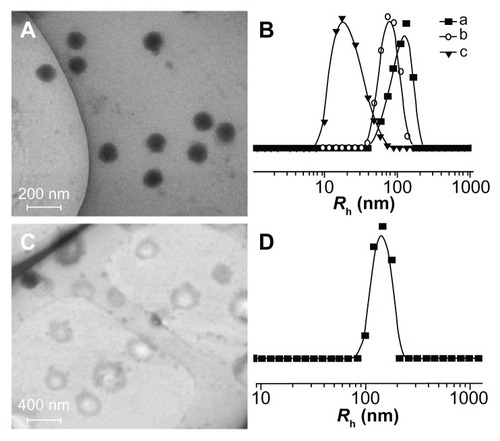
Figure 7 Release profiles of DOX from nanomedicines based on (A) (a1, a2, a3) mPEG-P(LA4-co-GA9)2, (b1, b2, b3) mPEG-P(LA12-co-GA9)2, and (c1, c2, c3) mPEG-P(LA45-co-GA15)2 micelles at pH (a1, b1, c1) 5.3, (a2, b2, c2) 6.8, and (a3, b3, c3) 7.4; (B) mPEG-P(LA24-co-GA8)2 vesicles at pH (a) 5.3, (b) 6.8, and (c) 7.4 in PB at 37°C.
Note: Data are presented as mean ± SD (n = 3).
Abbreviations: DOX, doxorubicin; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide); PB, phosphate buffer; SD, standard deviation.
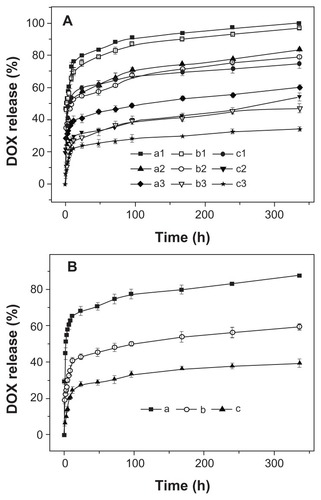
Figure 8 Representative CLSM images of HeLa cells incubated with free DOX and nanomedicines for 2 hours: incubated with (A) free DOX and nanomedicines based on (B) mPEG-P(LA4-co-GA9)2 micelle, (C) mPEG-P(LA12-co-GA9)2 micelle, (D) mPEG-P(LA24-co-GA8)2 vesicle, and (E) mPEG-P(LA45-co-GA15)2 micelle. For each panel, the images from left to right show cell nuclei stained by DAPI (blue) and cellular DOX fluorescence (red), and overlays of the two images.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DOX, doxorubicin; CLSM, confocal laser scanning microscopy; HeLa, Henrietta Lacks; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide).
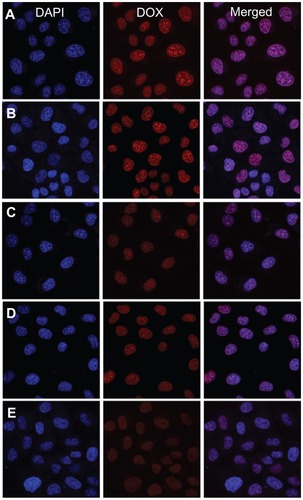
Figure 9 In vitro cytotoxicities of (a) PEI25K and nanoparticles from (b) mPEG-P(LA4-co-GA9)2, (c) mPEG-P(LA12-co-GA9)2, (d) mPEG-P(LA24-co-GA8)2, and (e) mPEG-P(LA45-co-GA15)2 toward HeLa cells.
Note: Data are presented as mean ± SD (n = 6).
Abbreviations: PEI, polyethylenimine; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide); HeLa, Henrietta Lacks; SD, standard deviation.
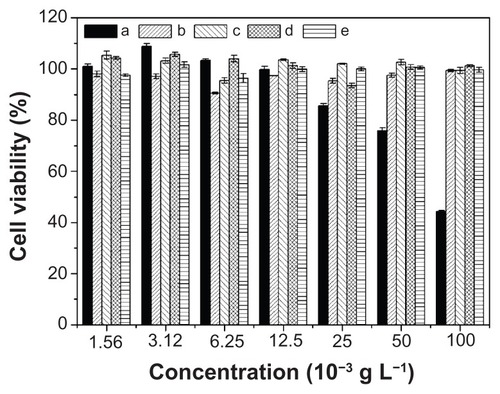
Figure 10 In vitro cytotoxicities of free DOX and nanomedicines based on (a) mPEG-P(LA4-co-GA9)2, (b) mPEG-P(LA12-co-GA9)2, (c) mPEG-P(LA24-co-GA8)2, and (d) mPEG-P(LA45-co-GA15)2 at (A) 24, (B) 48, and (C) 72 hours toward HeLa cells.
Note: Data are presented as mean ± SD (n = 6).
Abbreviations: DOX, doxorubicin; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide); HeLa, Henrietta Lacks; SD, standard deviation.
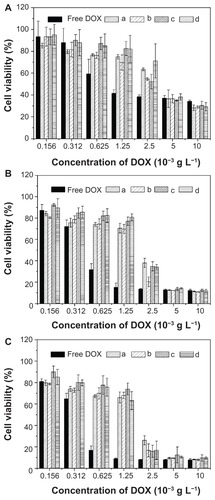
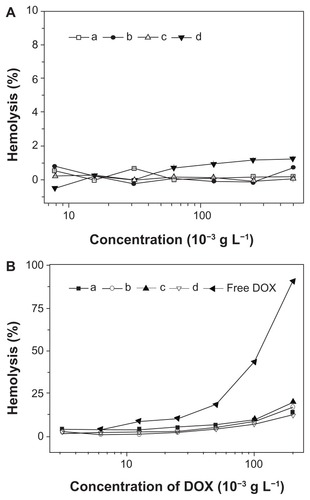
Figure 11 Percentage of RBC hemolysis incubated with (A) nanoparticles from (a) mPEG-P(LA4-co-GA9)2, (b) mPEG-P(LA12-co-GA9)2, (c) mPEG-P(LA24-co-GA8)2, and (d) mPEG-P(LA45-co-GA15)2, and (B) (a–d) homologous nanomedicines and free DOX. Physiological saline (−) and Triton X-100 (10 g L−1) (+) were used as negative and positive controls, respectively.
Note: Data are represented as mean ± SD (n = 3).
Abbreviations: DOX, doxorubicin; RBC, red blood cell; mPEG, monomethoxy poly(ethylene glycol); P(LA-co-GA), poly(L-lactide-co-glycolide); SD, standard deviation.