Figures & data
Figure 1 Schematic representation for preparation of RGD-DXRL-PEG.
Notes: The preformed liposomes were prepared by thin film hydration and polycarbonate membrane extrusion. Doxorubicin was loaded using a transmembrane ammonium sulfate gradient. cRGD was then covalently coupled to the liposomal surface via a chemical reaction between a thiol and maleimide group, followed by incubation with an aqueous micellar solution of DSPE-PEG2000.
Abbreviations: HSPC, hydrogenated soybean phosphatidylcholine; CHOL, cholesterol; MBPE, maleimidobenzoylphosphatidylethanolamine; DSPE-PEG2000, N-(carbonyl-methoxypolyethylene glycol 2000)-1, 2-distearoyl-sn-glycero-3-phosphoethanolamine sodium salt; DRUG, doxorubicin; DXRL-PEG, DXR-loaded PEGylated liposomes; RGD-DXRL-PEG, cRGD-modified DXRL-PEG.
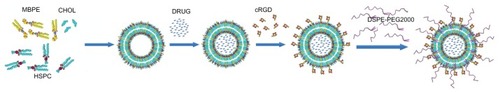
Figure 2 Size distribution of DXR-encapsulating liposomes determined by dynamic light scattering using a NICOMP 380 ZLS: size distribution of DXRL-PEG (A), and RGD-DXRL-PEG (B).
Abbreviations: DXR, doxorubicin; DXRL-PEG, DXR-loaded PEGylated liposomes; RGD-DXRL-PEG, cRGD-modified DXRL-PEG.
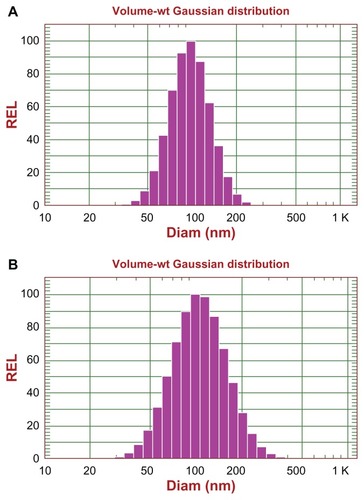
Figure 3 High-performance liquid chromatography of cRGD coupling with the liposomes. (A) Free cRGD (500 μg/mL) eluted with a retention time of approximately 10 minutes. (B) Excess free cRGD after coupling with the liposomes gave the peak for free cRGD. (C) The liposome sample following the coupling step showed no significant peak for free cRGD at around 10 minutes.
Abbreviations: DXR, doxorubicin; cRGD, cyclo(Arg-Gly-Asp-D-Phe-Cys).
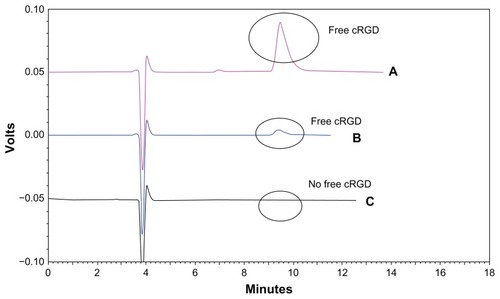
Figure 4 (A) Flow cytometry charts showing the cellular uptake of DXRL-PEG, RGD-DXRL-PEG, RGD-DXRL-PEG + 3 mM cRGD, and DXR solution by U87MG cells. (B) Mean fluorescence intensity as determined by flow cytometry experiments.
Notes: DXR solution served as a positive control while untreated cells were used as a negative control. U87MG cells were incubated with either free DXR, DXRL-PEG, or RGD-DXRL-PEG for 2 hours at 37°C. In the competitive binding experiment, excess free cRGD (3 mM) in culture medium was preincubated with U87MG cells for 20 minutes, followed by continued coincubation with RGD-DXRL-PEG for another 2 hours at 37°C. The cells were then washed with cold phosphate-buffered saline (pH 7.4) and cell-associated DXR was evaluated using flow cytometry. Data represent the mean ± standard deviation (n = 3). **P < 0.001.
Abbreviations: DXR, doxorubicin; DXRL-PEG, DXR-loaded PEGylated liposomes; RGD-DXRL-PEG, cRGD-modified DXRL-PEG.
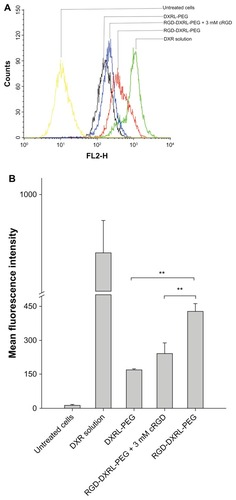
Table 1 Pharmacokinetic parameters of doxorubicin formulations in Sprague-Dawley rats following intravenous bolus administration
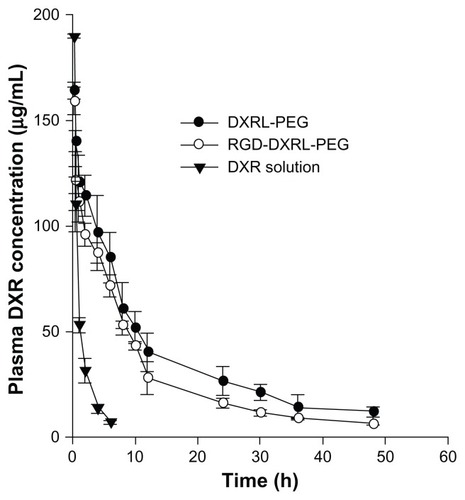
Figure 5 Plasma concentration versus time curves for DXR formulations in rats.
Notes: DXR formulations were administered via tail vein injection at a dose of 5 mg/kg. Each datum point is the average of 3–5 animals and the error bar is the standard deviation.
Abbreviations: DXR, doxorubicin; DXRL-PEG, DXR-loaded PEGylated liposomes; RGD-DXRL-PEG, cRGD-modified DXRL-PEG.