Figures & data
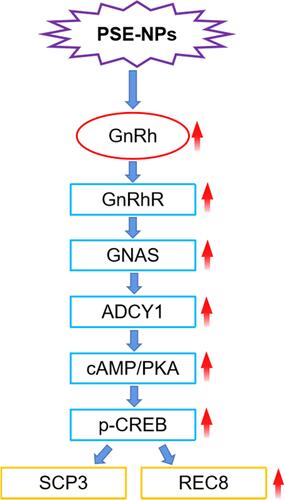
Table 1 Primer Sequences
Figure 1 Characterization of PSE-NPs: (A) SEM micrographs of the PSE NPs. (B) Dynamic light scattering of pseudoephedrine nanoparticles (PSE-NPs) and (C) in vitro release of PSE-NPs.
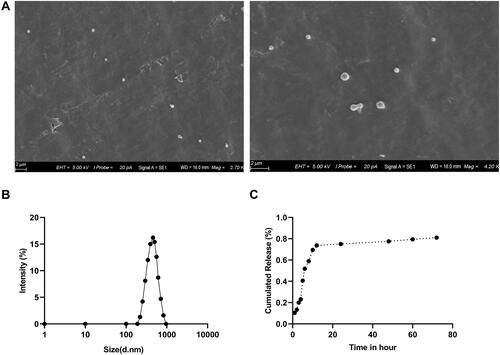
Figure 2 Effect of pseudoephedrine nanoparticles (PSE-NPs) on percent apoptosis of GC-1 cells and expression of apoptosis-related proteins: (A and B) Effect of PSE-NPs on percent apoptosis and survival of GC-1 cells in each group. (C) Effect of PSE-NPs on expression of apoptosis-related proteins of each group, scale bar 50 μm.
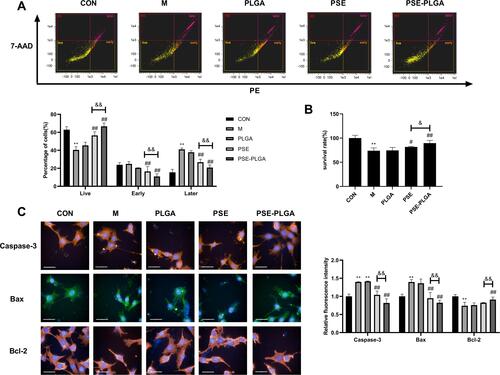
Figure 3 Effects of pseudoephedrine nanoparticles (PSE-NPs) on the mitochondrial functions and glycolysis rate of GC-1 cells: (A–C) Effects of PSE-NPs on basal respiration, ATP production, and maximal respiration in each group. (D and E) Effects of PSE-NPs on basal glycolysis and compensatory glycolysis in each group.
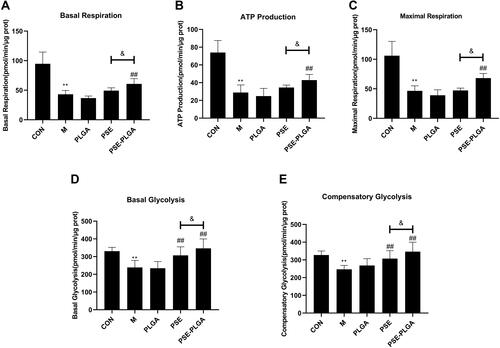
Figure 4 Effect of PSE-NPs on the GnRhR signaling pathway and expression of meiosis-related proteins SCP3 of GC-1 cells damaged by adriamycin: (A–E) Protein expression of GnRhR, GNAS, p-CREB and SCP3 in GC-1 cells, scale bar 50 μm.
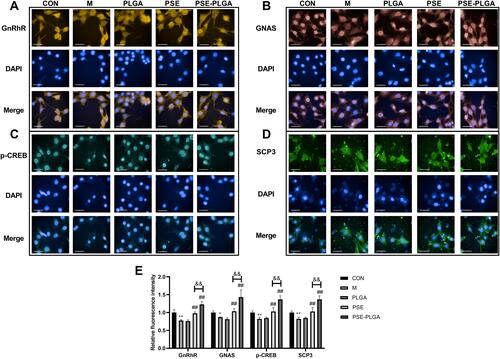
Figure 5 Effect of PSE-NPs on testicular histology in mice. The CON group showed a normal organizational structure. The Model group and PLGA group showed cell shedding and a decrease in the spermatogenic-cell layer. Compared with the Model group, the PSE group and PSE-PLGA group had significantly improved histology, and the PSE-PLGA group had the best effect. Black arrows mark showing spermatogenic cell layers.
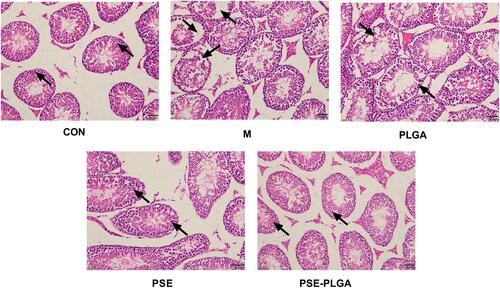
Table 2 Effects of PSE-NPs on Bodyweight, Testicular Indices, and Sperm Motility in Mice Treated with Adriamycin
Figure 6 Effects of PSE-NPs on percent apoptosis, oxidative stress, and expression of apoptosis-related proteins in mouse testicular cells: (A) Percent apoptosis in the testicular tissue of mice in each group. (B) mRNA expression of IL-1β, caspase 3 and TNF-α in the testicular tissue of mice each group measured by RT-qPCR. (C) Protein expression of Bax and Bcl-2 in the testicular tissue of mice in each group measured by Western blotting. (D) Levels of SOD and MDA in the testicular tissue of mice in each group.
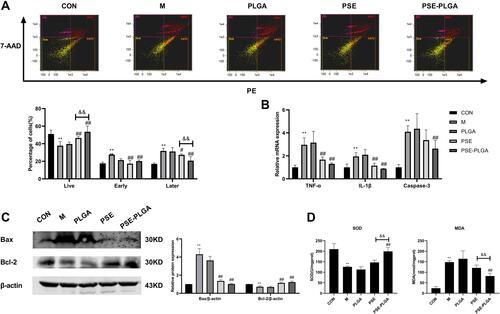
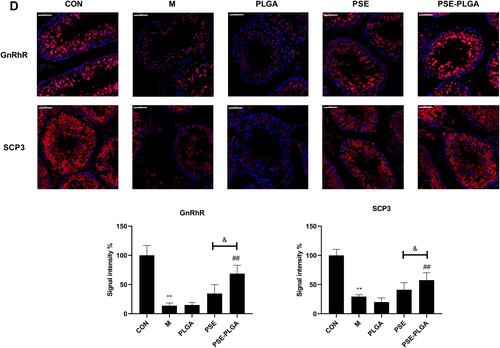
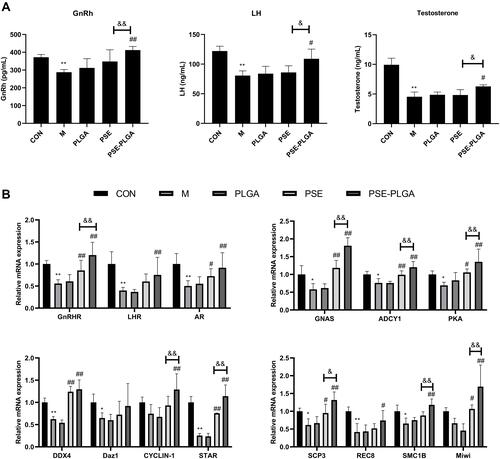
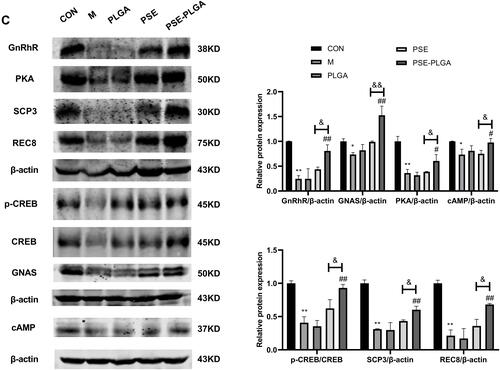
Figure 7 Effects of PSE-NPs on sex-hormone levels, GnRh signaling pathway-related proteins in testes, and indices of meiosis in mice: (A) Levels of GnRh, LH and testosterone in the serum of mice in each group. (B) mRNA expression of GnRh signaling pathway-related proteins and meiosis-associated genes in the testicular tissue of mice in each group was measured by RT-qPCR. (C) Protein expression of GnRh signaling pathway and meiosis-associated proteins in the testicular tissue of mice in each group was measured by western blotting. (D) The protein expression of GnRhR and SCP3 in the testicular tissue of mice in each group was measured by Immunofluorescence, scale bar 50 μm.