Figures & data
Table 1 Composition of rhG, Ch-I, and Ch-A
Figure 1 Synthetic scheme of (1) aCMG – (a) 1,6-hexamethylene diisocyanate, toluene, and pyridine at 80°C; and (b) rhG (targeting NH2 groups of Lys), triethylamine, and DMSO at 50°C, and (2) cCMG – (c) ethylenediamine and dichloromethane at RT; and (d) rhG (targeting COOH groups of Asp and Glu), using the EDC coupling agent, DMSO at RT.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin; DMSO, dimethyl sulfoxide; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride; rhG, recombinant human gelatin; RT, room temperature.
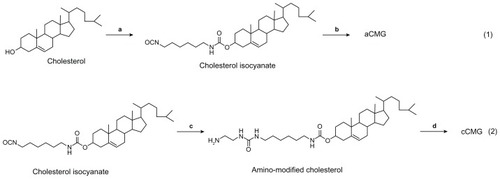
Figure 2 Illustration of the synthesis of aCMG and cCMG from rhG and their self-assembly in aqueous solution.
Notes: For aCMG, cholesterol isocyanate is conjugated with the amino groups of the lysine moieties of rhG and dispersed in aqueous solution to expose the free COO−groups, thereby attaining an overall negative charge. On the other hand, during the synthesis of cCMG, amino-modified cholesterol reacts with the COOH groups of aspartate and glutamate present in rhG. These disperse on hydration and expose free NH3+ groups to attain an overall positive charge.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin; rhG, recombinant human gelatin.
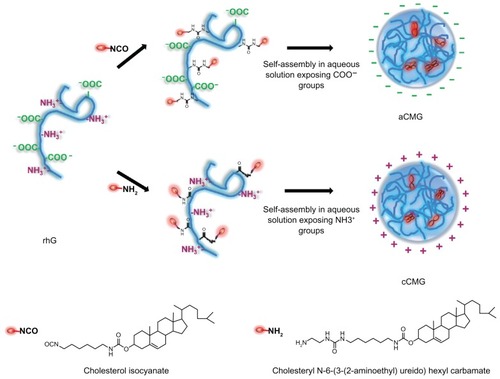
Figure 3 (A) MALDI-TOF mass spectra of the polymeric micelles, aCMG and cCMG. The masses of the cholesterol-modified gelatins were compared with that of native rhG to confirm the synthesis. (B) SEM images of aCMG and cCMG micelles. Samples were at a concentration of 1 mg/mL. Scale bar = 100 nm. The hydrodynamic size of the polymeric micelles estimated by DLS was larger than that observed by SEM. This discrepancy in size can be attributed to shrinkage of the particles during air-drying prior to SEM analysis. (C) Cytotoxicity of the polymeric micelles aCMG and cCMG versus native rhG (control) in DC 2.4 cells at concentrations of 10, 40, and 80 μg.
Notes: The cells were incubated with the polymers for 24 hours. Data are presented as the mean ± SD (n = 3); *P > 0.05.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin; DCs, dendritic cells; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; rhG, recombinant human gelatin; SD, standard deviation; SEM, scanning electron microscopy.
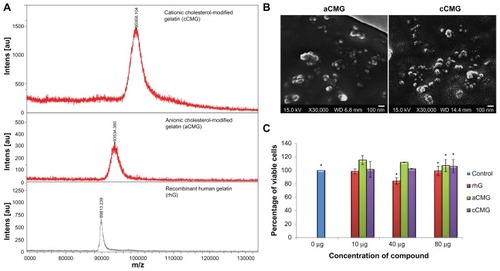
Figure 4 (A) FACS analysis of DC 2.4 cells incubated with rhG, aCMG and cCMG complexed with FITC-OVA for 1 hour at 37°C. Results are expressed as mean fluorescence intensity of the specific uptake of protein. This was assessed by subtracting the mean fluorescence intensity of cells incubated at 4°C from the value obtained at 37°C. (B) Fluorescent images show the cell uptake of FITC-OVA (green) in dendritic (DC 2.4) cells treated with FITC-OVA alone, or with rhG, aCMG, or cCMG at 37°C for 4 hours in RPMI-1640 medium supplemented with 10% fetal bovine serum.
Notes: After incubation, the cells were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde. Cells were stained with DAPI for nuclear DNA (blue). Scale bars = 20 μm.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin; DCs, dendritic cells; FACS, fluorescence-activated cell sorting; FITC-OVA, fluorescein isothiocyanate-labeled ovalbumin; rhG, recombinant human gelatin.
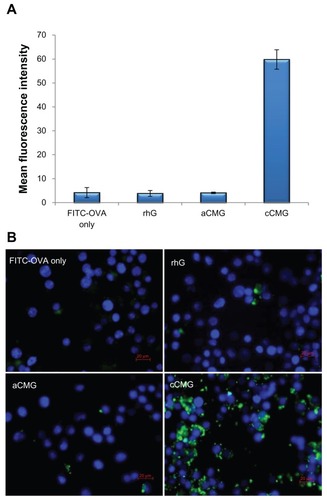
Figure 5 (A) Fluorescent images of DC 2.4 cells treated with rhG, aCMG, or cCMG complexed with FITC-OVA (green) in complete medium at 37°C for 5 hours. After 5 hours, the medium was replaced with fresh complete medium and incubated for an additional 19 hours. LysoTracker™ Red was added 20 minutes before fixing the cells with 4% paraformaldehyde to visualize acidic cell compartments: late endosomes and lysosomes (red). Cells were stained with DAPI for nuclear DNA (blue). Scale bars = 20 μm. (B) Processing of OVA was analyzed using OVA-DQ™.
Notes: DCs were incubated with OVA-DQ™ complexed with cCMG for 0, 5, and 24 hours and fixed. The figures are an overlay of green fluorescence (OVA-DQ™) and blue fluorescence (DAPI). Scale bars = 10 μm.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin; rhG, recombinant human gelatin; DC, dendritic cell; FITC-OVA, fluorescein isothiocyanate labeled-ovalbumin; DAPI, 4’,6-diamidino-2-phenylindole.
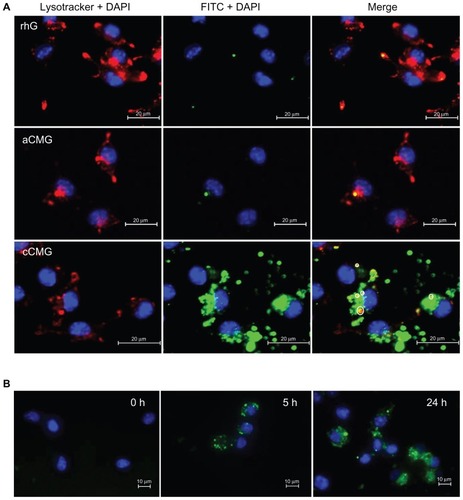
Figure 6 (A) Inhibition study by flow cytometry using various inhibitors of endocytosis. Results are expressed as the percentage of fluorescent positive cells. Control cells were treated with cCMG complexed with FITC-OVA only; **P < 0.01. (B) OVA-specific antibody titers after an intraperitoneal injection of free OVA and cCMG complexed with OVA in mice on day 7 after the first vaccination and day 21 after the first booster dose, by endpoint ELISA.
Notes: The results are expressed as the mean ± SD of three independent experiments. Significant differences between the groups: **P < 0.01.
Abbreviations: cCMG, cationic cholesterol-modified gelatin; FITC-OVA, fluorescein isothiocyanate-labeled ovalbumin; OVA, ovalbumin; rhG, recombinant human gelatin; SD, standard deviation.
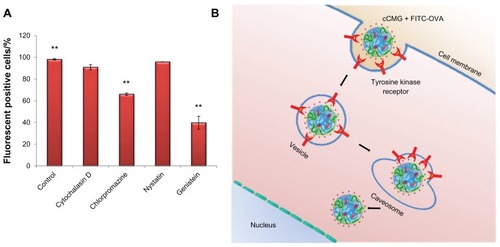
Figure 7 Specific antibody titers against OVA in murine blood were determined by ELISA after injection of PBS, OVA, or OVA-cCMG.
Notes: The results are expressed as the mean ± SD of three independent experiments. Significant differences between the groups: *P < 0.05; **P < 0.01.
Abbreviations: cCMG, cationic cholesterol-modified gelatin; OVA, ovalbumin; PBS, phosphate-buffered saline.
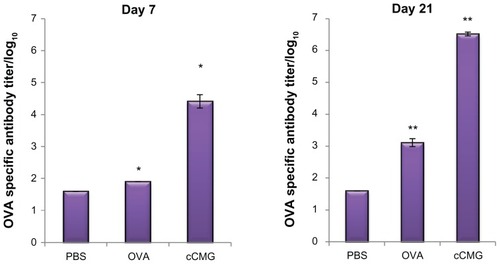
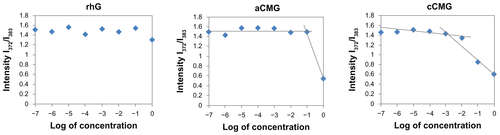
Figure S1 Critical micelle concentration (CMC) values of aCMG (1.7 × 10−2 mg/mL) and cCMG (2.0 × 10−3 mg/mL) analyzed by fluorescence spectrometry with the fluorescence intensity ratio I1/I3 of pyrene as a function of the concentration of modified gelatin.
Note: The native rhG does not form micelles and so does not have any CMC.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin.
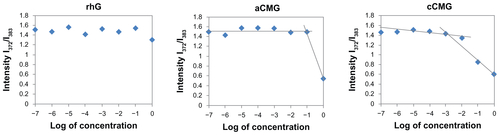
Figure S2 Confirmation of complex formation with OVA by aCMG or cCMG, as determined by native polyacrylamide gel electrophoresis analysis.
Notes: The gel was stained with Coomassie Brilliant Blue (stacking gel −3% and separating gel −12.5%). Lane 1, OVA; Lane 2, aCMG with OVA; Lane 3, cCMG with OVA.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin; OVA, ovalbumin.
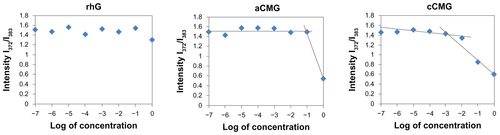
Figure S3 FACS analysis of the cellular uptake of free FITC-OVA, rhG, aCMG, and cCMG at 4 and 37°C.
Abbreviations: aCMG, anionic cholesterol-modified gelatin; cCMG, cationic cholesterol-modified gelatin; FACS, fluorescence-activated cell-sorting; FITC-OVA, fluorescein isothiocyanate-labeled ovalbumin; MFI; mean fluorescence intensity; rhG, recombinant human gelatin.
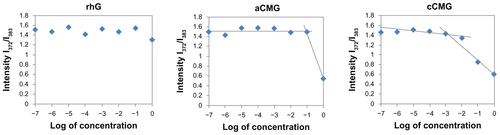
Figure S4 Fluorescence microscopy results of a time-course study on the uptake of cCMG by DC 2.4 cells after 2 and 24 hours.
Note: Intracellular FITC-OVA (green) was observed.
Abbreviations: cCMG, cationic cholesterol-modified gelatin; DCs, dendritic cells; FITC-OVA, fluorescein isothiocyanate-labeled ovalbumin.