Figures & data
Table 1 Synthesis and characterization of poly(amido amine)s
Figure 1 Synthetic scheme of dual-responsive poly(amido amine)s.
Abbreviations: CBA, N,N′-cystaminebisacrylamide; HDA, hexamethylene diamine; HMBA, hexmethylenebisacrylamide; KDA, [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea]; NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.
![Figure 1 Synthetic scheme of dual-responsive poly(amido amine)s.Abbreviations: CBA, N,N′-cystaminebisacrylamide; HDA, hexamethylene diamine; HMBA, hexmethylenebisacrylamide; KDA, [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea]; NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.](/cms/asset/1827a7e9-533f-4b91-a02d-de957336cb2e/dijn_a_37334_f0001_c.jpg)
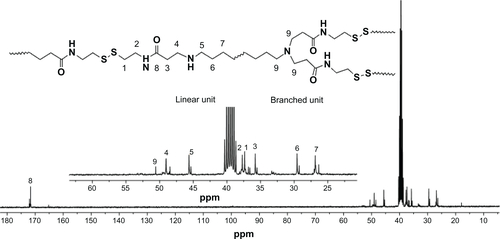
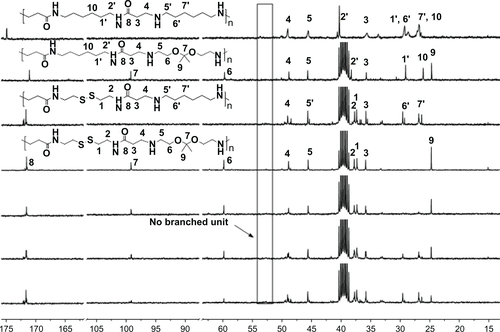
Figure 2 1H-nuclear magnetic resonance spectra of DualR1, DualR2, DualR3, DualR4, RedoxR, and AcidR in d6-dimethylsulfoxide, and NonR in D2O.
Abbreviations: NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.
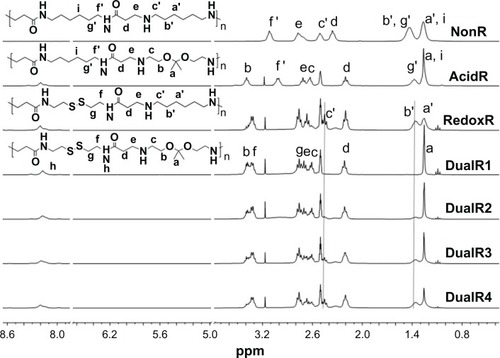
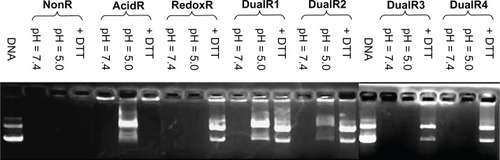
Figure 4 DNA condensation efficiency of various DNA/poly(amido amine) polyplexes at different N/P ratios.
Abbreviations: NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.
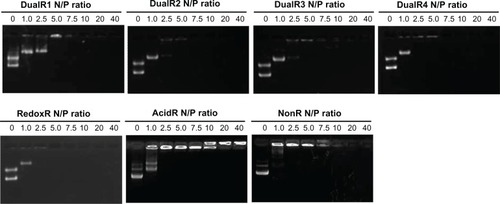
Figure 5 (A) Particle sizes and (B) zeta potential of various DNA/poly(amido amine) polyplexes at different N:P ratios.
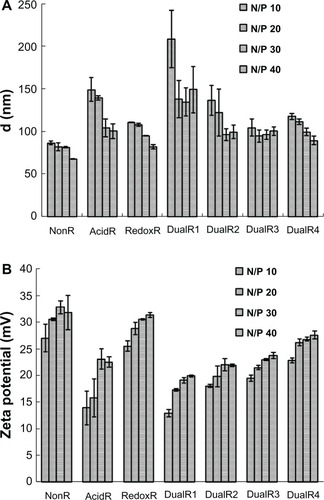
Table 2 Cytotoxicity of poly(amido amine)s in three cell lines
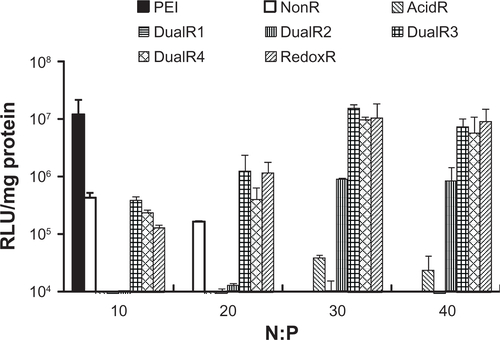
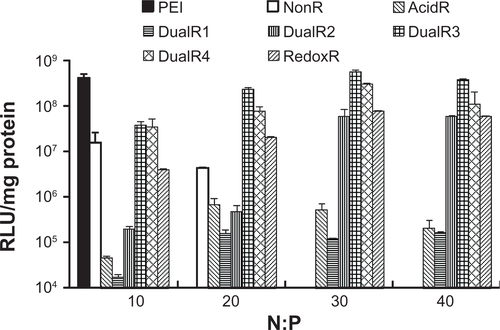
Figure 7 Luciferase expression of DNA/poly(amido amine) complexes in HePG2 cells.
Abbreviations: PEI, polyethylenimine; RLU, relative light units.
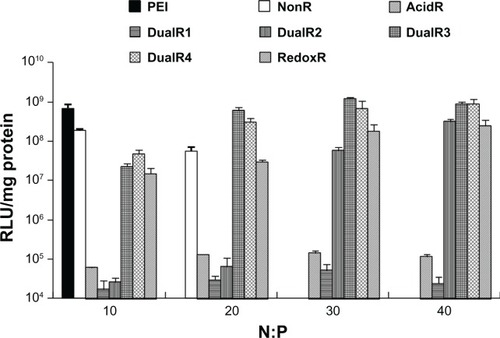
Figure 8 Fluorescence microscopy of green fluorescent protein transfection of DNA/poly(amido amine) polyplexes.
Abbreviation: PEI, polyethylenimine.
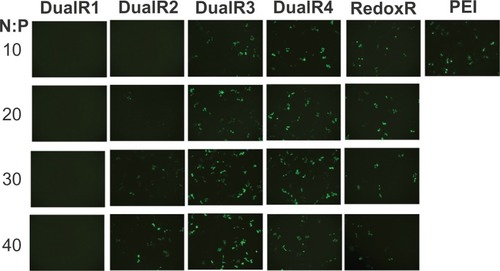
Figure 9 Green fluorescent protein expression of DNA/PAA polyplexes on flow cytometry.
Abbreviations: PAA, poly(amido amine)s; PEI, polyethylenimine.
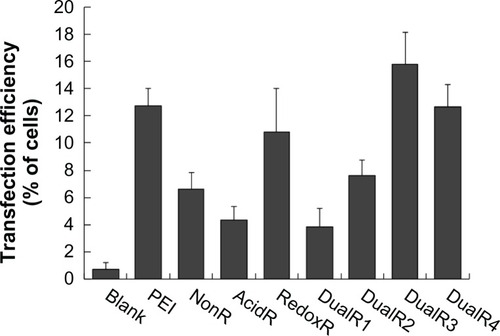
Figure S1 Synthetic scheme of [1,1′-(2,2′-(propane-2,2-diylbis(oxy) bis(ethane-2,1-diyl))diurea] (KDA).
![Figure S1 Synthetic scheme of [1,1′-(2,2′-(propane-2,2-diylbis(oxy) bis(ethane-2,1-diyl))diurea] (KDA).](/cms/asset/74267f00-94d4-45ef-918d-ae4598448c88/dijn_a_37334_sf0001_b.jpg)
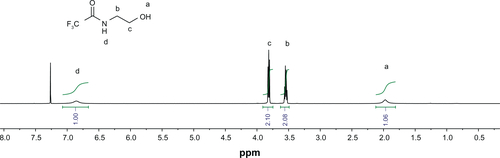
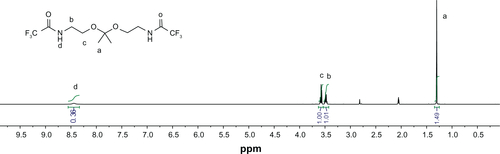
Figure S4 1H nuclear magnetic resonance spectrum of [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea] (KDA) compound in DCCl3.
![Figure S4 1H nuclear magnetic resonance spectrum of [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea] (KDA) compound in DCCl3.](/cms/asset/6d5472f0-9efa-490a-bf55-fee5c5459fcd/dijn_a_37334_sf0004_c.jpg)
Figure S5 1H nuclear magnetic resonance spectrum of N,N′-cystaminebisacrylamide (CBA) in d6-dimethylsulfoxide.
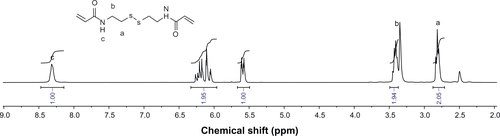
Figure S6 1H nuclear magnetic resonance spectrum of hexmethylenebisacrylamide (HMBA) in d6-dimethylsulfoxide.
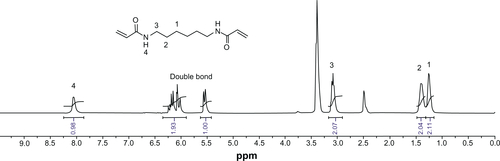
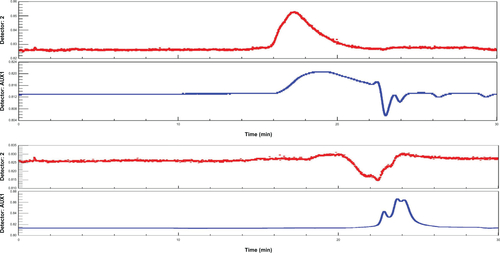
Figure S9 Gel permeation chromatography of DualR1 before (up) and after (down) incubation with 20 mM DTT.