Figures & data
Table 1 Enrichment of Phage Particles Against SARS-CoV-2 RBD-Specific Nanobodies During Three Rounds of Panning
Figure 1 Screening and recombinant expression of nanobodies. (A) Schematic diagram of the process of screening Nanobodies from a naïve alpaca VHH library. (B) Reactions from the 96 clones binding with SARS-CoV-2 RBD protein. (C) The phylogenetic tree of nanobodies with higher RBD reactions constructed using MEGA5.2 software. (D) Soluble prokaryotic expression and purification of four recombinant nanobodies. The destined proteins were indicated by the red arrows. (E) Alignment of the amino acid sequences of specific nanobodies with different amino acid sequences of CDR3.
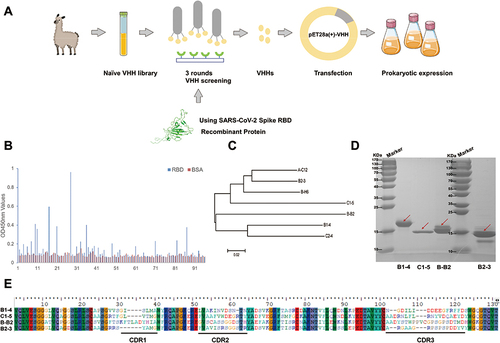
Table 2 The Results of Binding Affinity Between Nanobodies and RBD Using SPR
Table 3 Inhibition Rate Between Two Nanobodies
Figure 2 Affinity and binding sites of neutralizing nanobodies against RBD. (A) Structure docking model of nanobodies bound to RBD using DS. The antigen-binding epitopes are highlighted by yellow color. (B) RBD sequences of SARS-CoV-2 WT with a highlighted footprint of the four nanobodies (colored in light red). (C) The results of peptide-based ELISA of monomer nanobodies. (D) Exploring the RBD-ACE2 blocking activities (%inhibition) of four nanobodies to WT, Delta, and Omicron (B.1.1.529) RBD using sVNTs. (E) Measurement of the neutralization potency of nanobodies using pseudovirus neutralization assays.
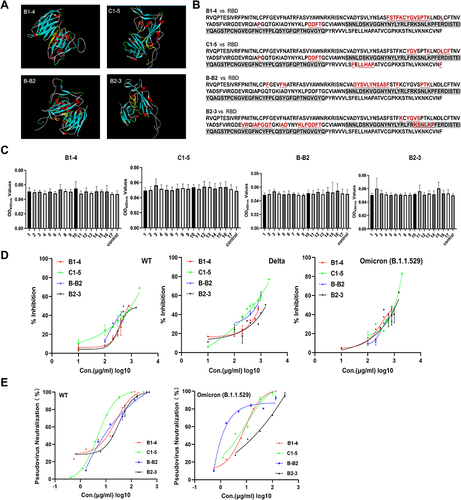
Table 4 The IC50 of Monovalent and Multivalent Nanobodies in the Pseudovirus Neutralization Assays
Figure 3 Conjugation and structural characterization of multivalent nanobodies. (A) Schematic diagram of multivalent nanobodies construction. (B) SDS-PAGE of the assembled products of AaLS and nanobodies at different molar ratios. (C) Negative-stain TEM images of the Spycatcher modified AaLS, Scale bar, 50 nm. Negative-stain TEM images of recombinant multivalent nanobodies, Scale bar, 50 nm.
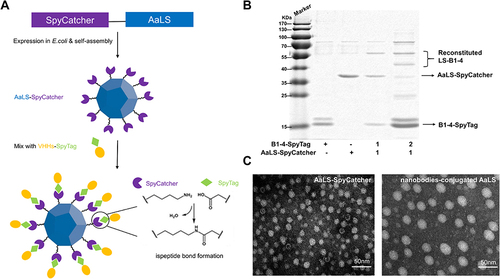
Figure 4 Comparison of the affinity and neutralizing activity of multivalent nanobodies. (A) Measurement of the binding ability of monovalent and recombinant multivalent nanobodies using indirect ELISA. (B) RBD-ACE2 blocking activities of monomer nanobodies and their multivalent pattern characterized using sVNTs. (C) Measurement of the neutralization potency of multivalent nanobodies using pseudovirus neutralization assays. (D) The epitopes recognized by multivalent nanobodies, groups that are statistically different from the control group are marked with * (E) Sequence comparison of SARS-CoV-2 variants RBD. The peptides 5 and 16 were highlighted in yellow.
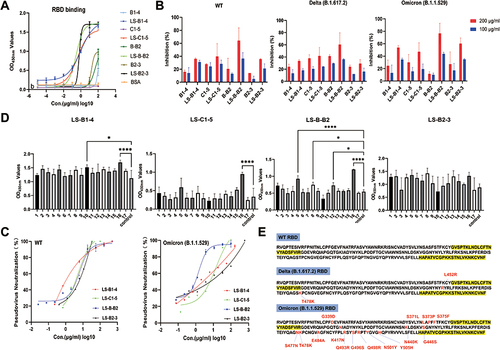
Table 5 17 Peptides of SARS-COV-2 WT RBD
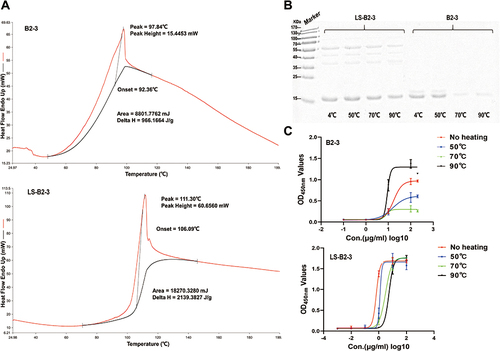
Figure 5 Nanobodies conjugated to the nanoplatform have good thermal stability. (A) DSC analysis of B2-3 and LS-B2-3. (B) SDS-PAGE of B2-3 and LS-B2-3 after heating at different temperatures. (C) Measurement of the binding ability of B2-3 and LS-B2-3 after heating at different temperatures using indirect ELISA.