Figures & data
Table 1 Primers Used in This Manuscript
Figure 1 Isolation and identification of M2-exos. (a). Representative pictures of THP-1 cells, M0 macrophages, and M2 polarized macrophages; (b). qRT-PCR analysis of expression of M0 polarized macrophage markers CD11b, CD14, and CD68; (c). qRT-PCR analysis of expression of M1 polarized macrophage marker iNOS and M2 polarized macrophage marker, IL-10, and CD206 -PCR analysis; (d). TEM observation of M2 polarized macrophage exosomes morphology (scale bar, 100 nm); (e). Nanoparticle particle size analysis showed that extracted exosomes were approximately 77.48 nm in diameter; (f). Western blotting detection of the expression of calnexin, HSP70, TSG101, and CD9 in exosomes of M0 polarized macrophages and M2 polarized macrophages and cell lysates; (g). Fluorescence microscopy of HemECs uptake of M2 exo (EvLINK labeled exosomes, CellLINK labeled cell membranes) (Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001; ns, nonsignificant).
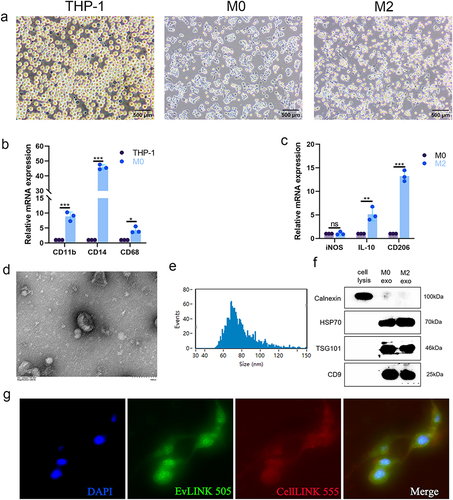
Figure 2 M2-exos regulates biological functions of HemECs. (a). Cell proliferation capacity assessed by CCK-8 assay; (b and c). Wound healing assay used to examine migration rate of HemECs; (d and e). Invasive capacity of HemECs assessed using Transwell chambers, and the number of cells passing through the chamber membrane were counted; (f–i). Tube formation assay representative images and number of junctions and meshes and total segments length; (j). Relative expression analysis of MIR4435-2HG in M0/M2 macrophage-derived exosomes using qRT-PCR (Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001).
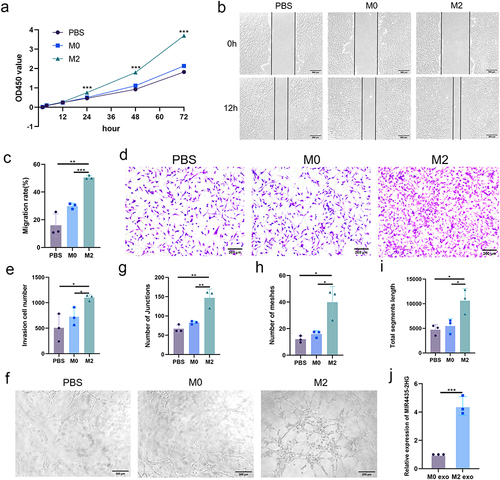
Table 2 Baseline Characteristics of Patients with IHs and Control Subjects
Figure 3 MIR4435-2HG is elevated in IHs and regulates the biological functions of HemECs in vivo. (a). Detection of MIR4435-2HG expression and localization in IH and normal tissues using FISH. Scale bar, 50 μm.; (b). qRT-PCR analysis of MIR4435-2HG expression in IHs and normal tissues; (c). Detection of subcellular localization of MIR4435-2HG in HemECs using FISH. Scale bar, 20 μm.; (d). Detection of knockdown and overexpression efficiency of MIR4435-2HG using qRT-PCR; (e). Assessment of cell proliferation capacity using CCK-8 assay; (f and g). Wound healing assay was used to examine the migration rate of HemECs; (h and i). Use of Transwell chambers to assess invasive ability of HemECs and count the number of cells passing through the chamber’s membrane; (j–m). Representative images of tube formation assays and statistics of the number of junctions and meshes and total segments length (Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001).
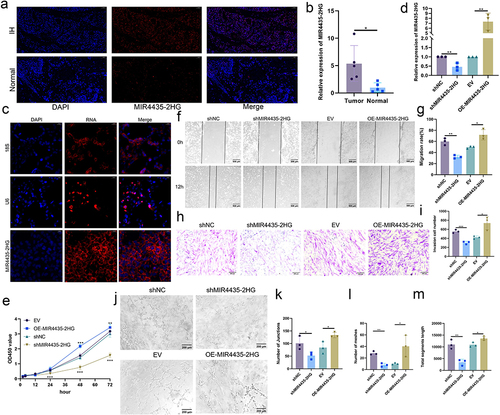
Figure 4 MIR4435-2HG downregulation inhibits the growth of HemECs in vivo. (a). Tumors of nude mice were dissected 2 weeks after subcutaneous injection in shNC and shMIR4435-2HG groups (5 mice in each group). (b). Tumor volume was significantly reduced in the shMIR4435-2HG group compared with that in the shNC group. (c). Tumor weight was significantly reduced in the shNC group compared with the shMIR4435-2HG group. (d). Typical H&E staining of tumors with abundant microvessels visible in the shNC group. The arrows indicate the microvessels. (e). Expression and localization of MIR4435-2HG in tumor tissues of shNC and shMIR4435-2HG groups were detected using FISH. Scale bar, 100 μm.(Student’s t-test, ***P < 0.001).
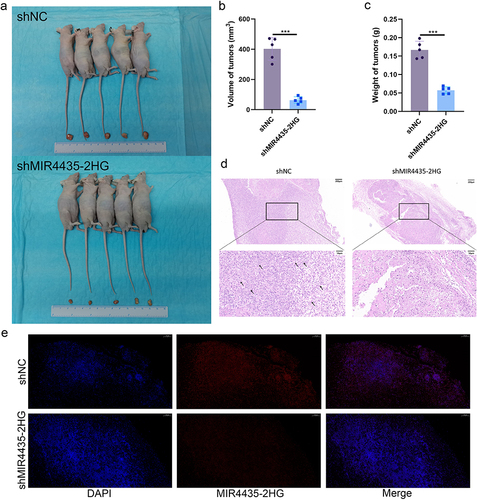
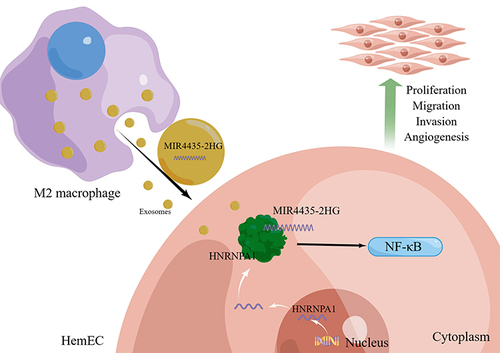
Figure 5 M2-exos regulates the biological functions of HemECs by transferring MIR4435-2HG. (a). qRT-PCR to detect MIR4435-2HG expression after addition of M2-exos; (b). Cell proliferation capacity assessed using CCK-8 assay; (c and d). Wound healing assay used to examine the migration rate of HemECs; (e and f). Transwell chambers were used to assess invasive ability of HemECs and count the number of cells passing through the chambers membrane. (g–j). Representative images of angiogenesis assays and statistics of junction number, mesh number, and total section length (Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001).
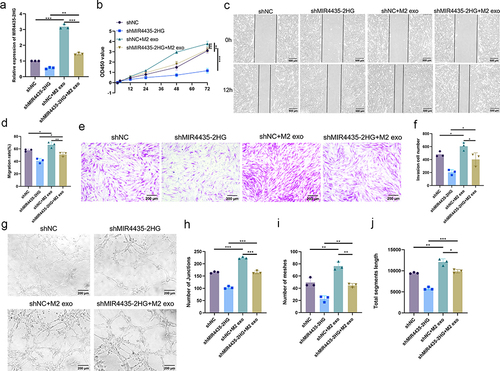
Figure 6 RNA-seq analysis of differential genes and regulatory pathways and MIR4435-2HG binds HNRNPA1 protein. (a). Cluster analysis of shMIR4435-2HG and shNC; (b). Volcano plot showing differentially expressed genes in shMIR4435-2HG and shNC (|log2FC| ≥ 1 and p-adjust < 0.05); (c). GSEA for NF-κB pathway; (d). Representative images (left) and histograms (right) of Western blotting of NF-κB signaling pathway NF-κB p65, p-p65, IκBα, and p-IκBα expression levels in HemECs after MIR4435-2HG knockdown; (e). MIR4435-2HG overexpression in representative images (left) and histograms (right) of Western blotting of NF-κB signaling pathway NF-κB p65, p-p65, IκBα, and p-IκBα expression levels in HemECs; (f). Mass spectrometry of protein obtained by RNA pull-down and HNRNPA1 was identified as MIR4435-2HG binding protein; (g). Western blotting to detect HNRNPA1 binding to MIR4435-2HG; (h). RIP showing HNRNPA1 binding to MIR4435-2HG; (i). Detection of knockdown or overexpression of HNRNPA1 expression in HemECs with MIR4435-2HG using qRT-PCR; (j). Representative images (left) and histograms (right) of Western blotting of HNRNPA1 expression levels in HemECs after MIR4435-2HG knockdown; (k). Western blotting of HNRNPA1 expression in HemECs after MIR4435-2HG overexpression representative images (left) and histograms (right) (Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001).
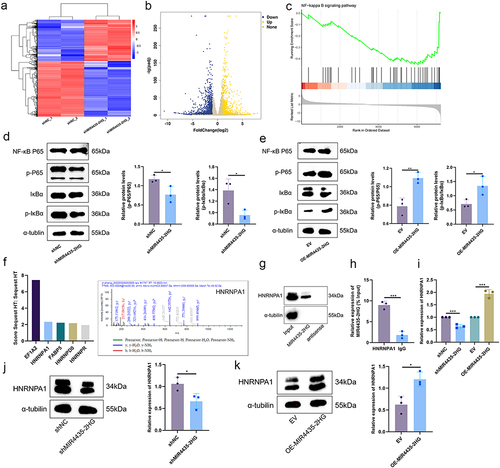
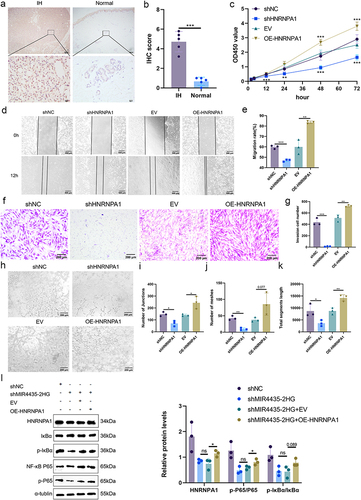
Figure 7 HNRNPA1 expression is elevated in IHs and regulates biological function of HemECs in vivo. (a and b). IHC staining and scoring of HNRNPA1 in IHs and normal tissues; (c). Cell proliferation capacity assessed by CCK-8 assay; (d and e). Wound healing assay used to examine the migration rate of HemECs; (f and g). Use of Transwell chambers to assess invasive capacity of HemECs and to count the number of cells passing through the chambers membrane; (h–k). Representative images of tube formation assays and statistics of the number of junctions and meshes and total segments length; (l). Representative images (left) and histograms (right) of Western blotting of HNRNPA1 expression and NF-κB signaling pathway molecules in HemECs (Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001, ns, nonsignificant).