Figures & data
Figure 1 Proposed reaction scheme for synthesis of OCMPEI copolymer.
Notes: LMWSC was modified to OCMCh (A) by monochloroacetic acid. OCMPEI copolymers (B) were synthesized by a chemically coupling reaction between EDC/NHS-activated carboxyl groups of OCMCh and amine groups of bPEI.
Abbreviations: bPEI, branched polyethylenimine; DMF, dimethylformamide; EDC, 1-ethyl-3-(30dimethylaminopropyl) carbodiimide hydrochloride; LMWSC, low molecular weight water soluble chitosan; NHS, N-hydroxysuccinimide; OCMCh, O-carboxymethyl chitosan; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine.
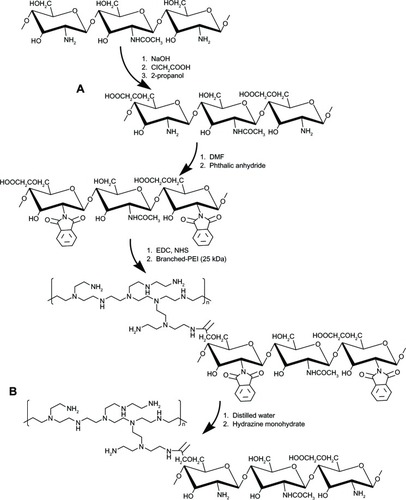
Figure 2 1H-NMR of OCMCh (A), bPEI (B), and OCMPEI (C).
Note: Numbers 1–7 and letters a–f indicate peaks of OCMCh and PEI, respectively.
Abbreviations: bPEI, branched polyethylenimine; NMR, nuclear magnetic resonance; OCMCh, O-carboxymethyl chitosan; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine.
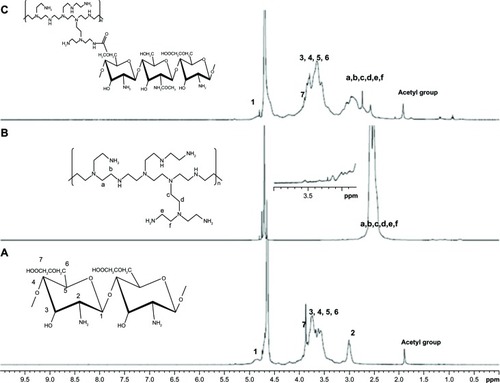
Figure 3 Gel retardation assay of the pDNA/OCMPEI polyplexes.
Notes: (A) Electrophoretic mobility shift assay of pDNA/OCMPEI polyplexes at weight ratios. (B) DNase protection assay of pDNA/OCMPEI polyplexes (w/w: 1:1 and 1:2) when treated with DNase I. (C) DNA-releasing assay from pDNA/OCMPEI polyplexes when treated with heparin.
Abbreviations: DNA, deoxyribonucleic acid; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; pDNA, plasmid DNA.
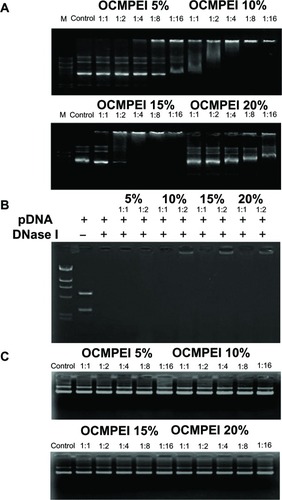
Figure 4 Particle size and morphological observation of pDNA/OCMPEI polyplexes.
Notes: (A) pDNA was complexed with OCMPEI 5% (1), 10% (2), 15% (3), and 20% (4) at a w/w ratio of 16 and analyzed by DLS. (B) spherical forms of pDNA/OCMPEI polyplexes were observed by using transmission electron microscope; (1–4) are the same percentages as in (A).
Abbreviations: DLS, dynamic light scattering; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; pDNA, plasmid deoxyribonucleic acid.
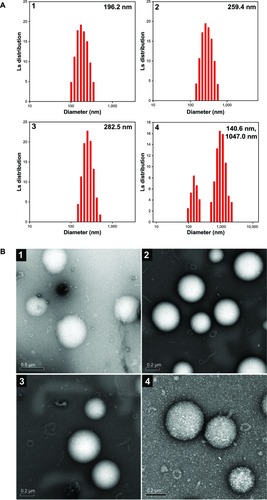
Figure 5 Cytotoxicity of copolymers and hemolytic effect of polyplexes with pDNA against HEK293 (A) and human erythrocytes (B), respectively.
Abbreviations: bPEI, branched polyethylenimine; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; pDNA, plasmid deoxyribonucleic acid.
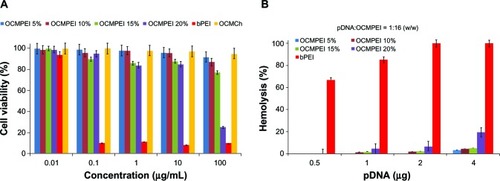
Figure 6 Gene transfection efficiency of pDNA/OCMPEI polyplexes in various cells.
Notes: (A) pEGFP gene transfection of polyplexes at a w/w ratio of 16, free OCMCh, free bPEI, and lipofectamine in HCT119 cells. (B) luciferase expression of pGL3 DNA complexes with OCMPEI copolymers, free OCMCh, free bPEI, and lipofectamine in HEK293, HCT119, and lovo cells.
Abbreviations: bPEI, branched polyethylenimine; DNA, deoxyribonucleic acid; OCMCh, O-carboxymethyl chitosan; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; pEGFP, plasmid DNA encoding green fluorescent protein.
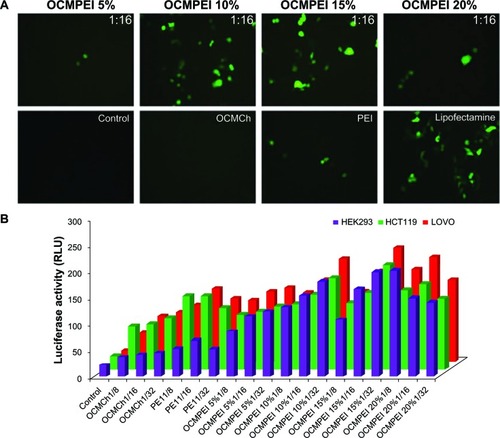
Figure 7 Gel retardation assay and morphological observation of siRNA/OCMPEI polyplexes.
Notes: (A) Electrophoretic mobility of siRNA complexed by (1) OCMPEI 5%, (2) 10%, (3) 15%, and (4) 20% at w/w ratios of 1:1∼1:16. (B) morphological observation of siRNA/OCMPEI 10% and 15% at a w/w ratio of 1:8 using TEM.
Abbreviations: OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; siRNA, small interfering ribonucleic acid; TEM, transmission electron microscopy.
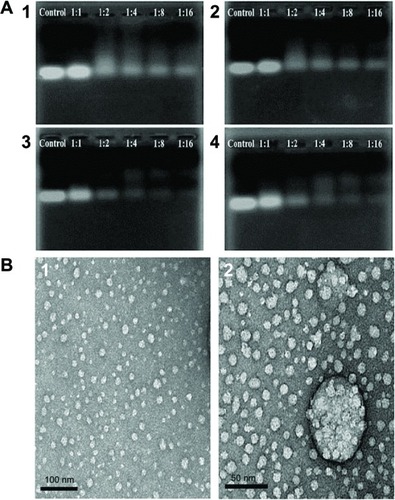
Figure 8 Gene silencing efficiency of siRNA/OCMPEI 10% and 15% polyplexes in HCT 119 cells.
Notes: (A) HCT 119 cells pretreated with pEGFP/OCMPEI were transfected with scrambled siRNA (600 ng) or GFP-siRNA (300 ng or 600 ng)/OCMPEI 10% and 15%. After 2 days, GFP gene silencing was observed by fluorescence microscope. (B) Western blotting analysis of GFP downregulation. Lane 1: control without pEGFP; lane 2: pretreated with pEGFP/OCMPEI 10%; lane 3: scrambled siRNA/OCMPEI 10%; lane 4: GFP-siRNA/OCMPEI 10% (300 ng); lane 5: GFP-siRNA/OCMPEI 10% (600 ng); lane 6: pretreated with pEGFP/OCMPEI 15%; lane 7: scrambled siRNA/OCMPEI 15%; lane 8: GFP-siRNA/OCMPEI 15% (600 ng), lane 9: GFP-siRNA/OCMPEI 15% (600 ng).
Abbreviations: OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; siRNA, small interfering ribonucleic acid; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; pEGFP, plasmid DNA encoding green fluorescent protein; GFP, green fluorescent protein.
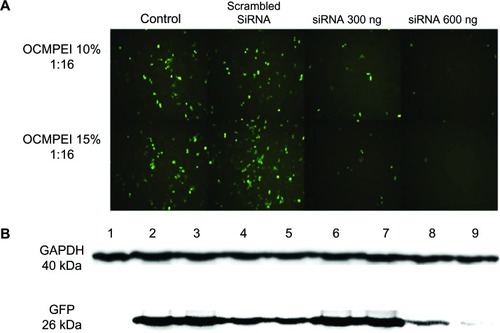
Figure 9 In vivo gene delivery of intravenous injected pEGFP/OCMPEI polyplexes to an ICR mouse.
Notes: OCMPEI 15% complexed with 30 μg of pEGFP at a w/w of 32:1 was injected through the mouse tail vein; after 5 days, the whole body (A) and vital organs (B) were fluorescently imaged.
Abbreviations: OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; pEGFP, plasmid DNA encoding green fluorescent protein.
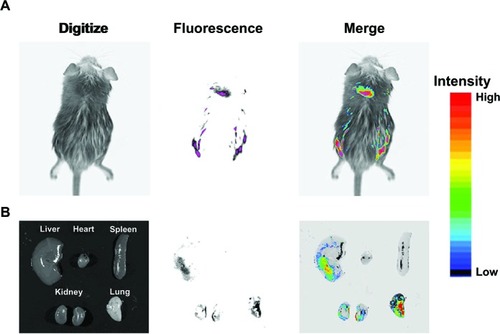
Figure 10 Effect of endocytic inhibitors on cellular uptake in HCT119 cells treated with pDNA/OCMPEI polyplexes.
Notes: (A) After 1-hour preincubation of endocytic inhibitors (3 μg/mL sodium azide, 10 μg/mL chlorpromazine, 8 μg/mL colchicine, and 6 μg/mL indomethacin) with HCT119 cells, pEGFP/OCMPEI 10% was transfected, after which the cell was analyzed by FACSCalibur flow cytometer. (B) The results were visualized using fluorescence microscopy.
Abbreviations: FITC-A, fluorescein isothiocyanate A; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; pDNA, plasmid deoxyribonucleic acid.
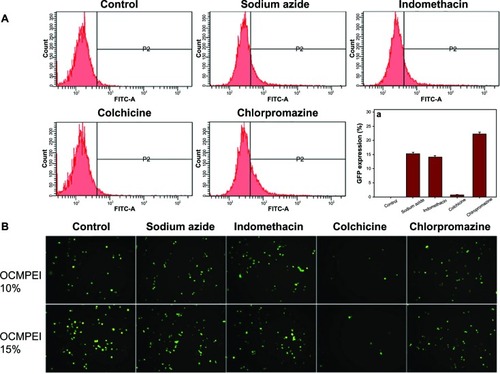
Figure S1 1H NMR of OCMPEI 5%, 10%, 15%, and 15% in D2O solvent.
Note: The red arrow indicates the increasing intensity peak of PEI.
Abbreviations: D2O, deuterium oxide; 1H NMR, 1H nuclear magnetic resonance; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine.
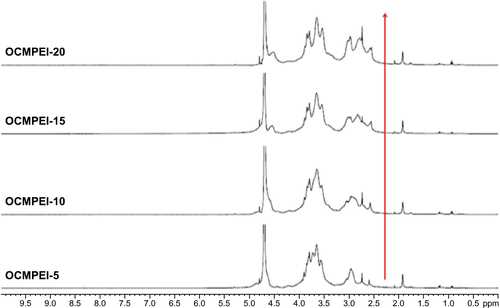
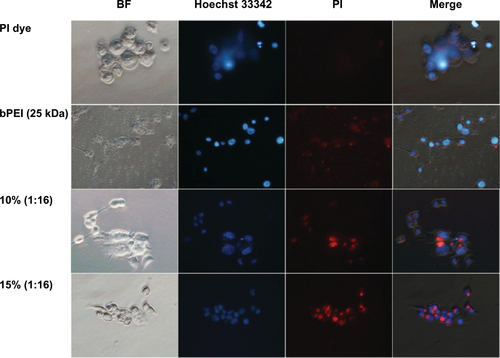
Figure S2 Cellular uptake of PI-labeled siRNA/OCMPEI polyplexes in HCT 119 cells.
Note: PI-labeled siRNA was complexed with OCMPEI 10% and 15% at a w/w ratio of 1:16 and cell nuclei were stained by Hoechst 33342.
Abbreviations: BF, bright field; OCMPEI, O-carboxymethyl chitosan-graft-branched polyethylenimine; PI, propidium iodide; siRNA, small interfering ribonucleic acid; bPEI, branched polyethylenimine.