Figures & data
Table 1 The most popular cross-linker reagents for coupling protein to nanoparticle based on their respective functions
Figure 1a The introduction of sulfhydryl groups by: 1) the reduction of protein disulfide bonds using reductive agents such as dithiotreitol (DTT = Cleland’s reagent). 2) Coupling protein primary amino groups with 2-iminothiolane (Traut; s reagent).
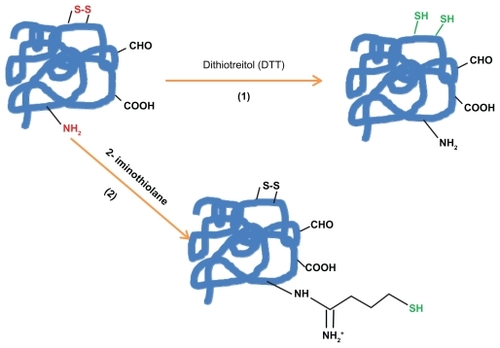
Figure 1b The introduction of sulfhydryl groups by: 3) Quenching of reactive protein aldehyde residues with cystaminiumdichloride reagents or 4) coupling of cystaminiumdichloride to carboxyl groups via 1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimide (EDC); both cases followed by the disulfide bonds reduction with DTT.
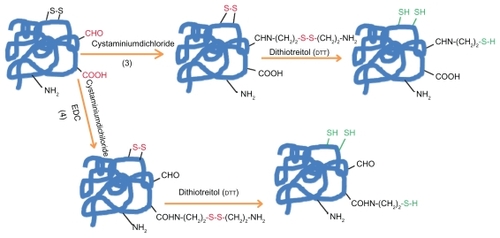
Figure 2 Protein–nanoparticle interactions: main factors that can affect proteins resulting in their denaturation. In this example, proteins are conjugated on amino functionalized nanoparticles using the cross-linker SPMB.
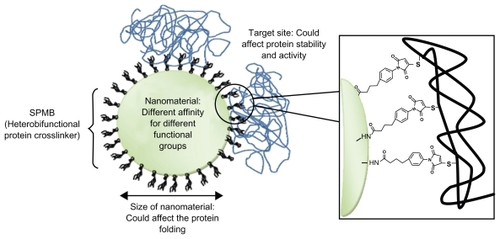
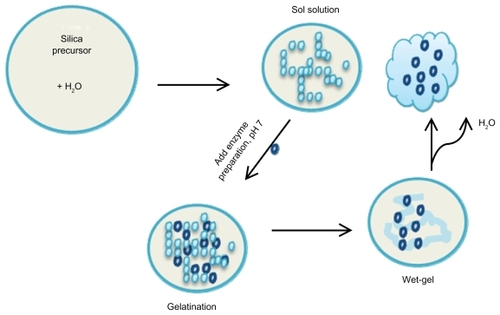
Figure 3 Entrapment of enzymes using sol-gel chemistry: A schematic overview of the sol-gel process. Several silicate precursors can be used to modify the surface chemistry of the sol-gels such as TMSO, APTES, MTMOS, and ETMOS.
Abbreviations: TMSO, tetramethyl orthosilicate; APTES, 3-aminopropyltriethoxysilane; MTMOS, methyltrimethoxysilane; ETMOS, ethyltrimethoxysilane.