Figures & data
Figure 1 CRPPR-R9/lipofectamine/mRNA complex can deliver GMT mRNAs to cardiac fibroblasts and induct direct reprogramming towards cardiomyocyte-like cells.
Notes: C-Lipo composed of a heart-targeting peptide (CRPPR-R9) and lipofectamine facilitates mRNA delivery in cardiac fibroblasts. GMT mRNA transfection induces partial direct reprogramming of cardiac fibroblasts towards cardiomyocytes.
Abbreviation: GMT, Gata4, Mef2c, and Tbx5.
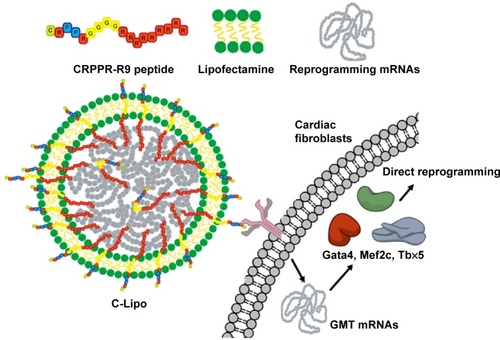
Figure 2 CRPPR-R9 facilitates mRNA delivery in cardiac fibroblasts and CRPPR-R9/lipofectamine improves mRNA transfection efficiency.
Notes: (A) Confocal images of cardiac fibroblasts treated with Alexa594-labeled mRNA/CRPPR-R9 complexes. mRNA and mRNA/CRPPR-R9 complexes were transfected into cardiac fibroblasts for 30 minutes. CRPPR-R9 induces facilitated mRNA uptake in cardiac fibroblasts. Alexa594-labeled mRNA and nucleus are shown in yellow and blue, respectively. Scale bar is 10 μm. (B) Transfection efficiency of eGFP mRNA on cardiac fibroblasts transfected by various combinations of CRPPR-R9 and lipofectamine and quantified by flow cytometry. CRPPR-R9 with lipofectamine increases eGFP mRNA transfection efficiency. (C) Mean eGFP fluorescence of cardiac fibroblasts quantified by flow cytometry. Mean eGFP fluorescence increases as transfection efficiency increases in CRPPR-R9 and lipofectamine transfection. The results represent the mean ± SE, n=4. *P<0.05; **P<0.01. (D) Images of eGFP fluorescence in cardiac fibroblasts with eGFP mRNA. Scale bar is 10 μm.
Abbreviations: eGFP, enhanced green fluorescence protein; SE, standard error.
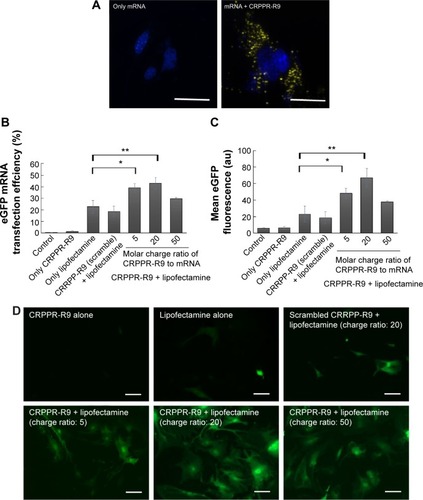
Figure 3 C-Lipo with reduced lipofectamine amount transfects cardiac fibroblasts efficiently without significant toxicity.
Notes: (A) Cell viability of C-Lipo (0.5 μL of lipofectamine with 7 μg of CRPPR-R9 peptide per 1 μg mRNA) after daily transfections. Cardiac fibroblasts were transfected with C-Lipo and Lipo daily for 1 week and cell viabilities were measured on days 4 and 7. C-Lipo allows mRNA transfection in cardiac fibroblasts without significant toxicity for 1 week compared to Lipo, which causes significant toxicity. (B) and (C) Flow cytometry analyses of cardiac fibroblasts treated with C-Lipo and Lipo. C-Lipo shows comparable levels of transfection efficiency and higher mean eGFP fluorescence compared to Lipo. The results represent the mean ± SE, n=5. *P<0.05; **P<0.01.
Abbreviations: eGFP, enhanced green fluorescence protein; SE, standard error; ns, not significant.
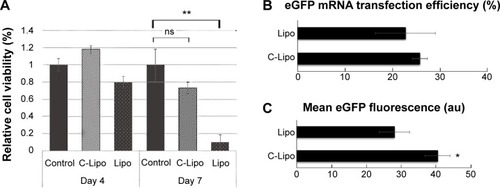
Figure 4 Transfection of GMT mRNAs on cardiac fibroblasts induces expression of cardiomyocyte marker genes and striated cardiac muscle structure.
Notes: (A) Fluorescence images of α-MHC promoter-driven GFP expression from GMT mRNA transfected cardiac fibroblasts. α-MHC-GFP cardiac fibroblasts were transfected daily with GMT mRNAs for 7 days and observed with fluorescence microscope. (a, b) Both bright field and green fluorescence images were taken with 10× magnification. (c, d) 20× magnification images were taken from two additional regions. Scale bar is 10 μm. (B) Immunohistochemistry staining of α-actinin in GMT mRNA/C-Lipo transfected cardiac fibroblasts. Inducted cardiomyocyte-like cells express α-actinin and have striated cardiac muscle structure. Scale bar is 2 μm.
Abbreviations: GFP, green fluorescence protein; GMT, Gata4, Mef2c, and Tbx5.
Figure 5 Transfection of GMT mRNAs via C-Lipo induces expression of cardiomyocyte marker genes from transfected cardiac fibroblasts.
Notes: Quantification of the relative expression of cardiomyocyte marker genes after 2 weeks of transfection. GMT mRNA transfection via C-Lipo drives cardiac fibroblasts to express cardiomyocyte marker genes: Actc1, Actn2, Gja1, Hand2, and Tnnt2. The results represent the mean ± SE, n=3. *P<0.05; **P<0.01.
Abbreviations: GMT, Gata4, Mef2c, and Tbx5; SE, standard error.
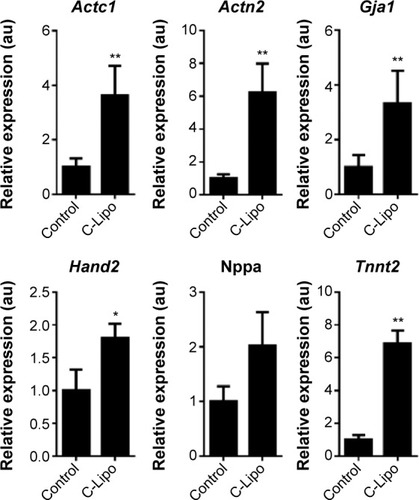
Figure 6 Stoichiometric ratio of GMT mRNAs influences cardiomyocyte marker gene expression patterns.
Notes: Quantification of the relative expression of cardiomyocyte marker genes in cardiac fibroblasts transfected with various stoichiometric ratios of GMT mRNAs. Gene expression levels of Actc1, Actn2, Gja1, Hand2, Nppa, and Tnnt2 were affected by the stoichiometric ratio of GMT mRNAs. The results represent the mean ± SE, n=3. *P<0.05; **P<0.01; statistical significance was tested between the control and each sample.
Abbreviations: GMT, Gata4, Mef2c, and Tbx5; SE, standard error.
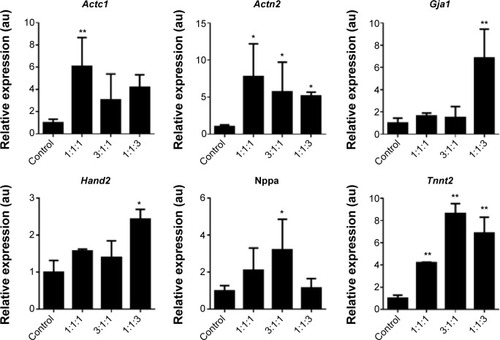
Figure S1 eGFP mRNA transfection was optimized with various transfection reagents.
Notes: Various ratios of eGFP mRNA and transfection reagents were tested for cardiac fibroblast transfection. Compared to mouse embryonic fibroblasts that are easy to transfect with lipofectamine, cardiac fibroblasts show inefficient transfection in tested conditions. Among the tested transfection reagents, lipofectamine (1 μg mRNA to 2 μL lipofectamine) shows the best transfection efficiency. Fluorescence images were taken from a confluent 24-well plate. Scale of width of each image is 50 μm. Fluorescence images of transfection reagents that had very inefficient transfection including TransIT-X2, TransIT-mRNA (Mirus Bio LLC, Madison, WI, USA), bPEI, and PEG-polylysine (22 ethylene glycol units, 50 lysine units) are not shown. Transfection was conducted as described in the main text.
Abbreviations: bPEI, branched polyethyleneimine; eGFP, enhanced green fluorescence protein.
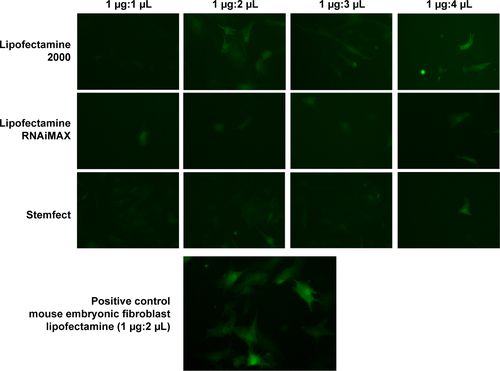
Figure S2 Electrophoretic mobility shift assay shows complexation of mRNA and CRPPR-R9.
Notes: EMSA was conducted to investigate if CRPPR-R9 complexes with mRNA. A mixture of CRPPR-R9 and mRNA produced visible retarded RNA bands from a molar charge ratio of one ([positive amines in peptide] to [phosphates in mRNA]) and a complete complexation was observed in a molar charge ratio of 4 or above. Unbound mRNA bands look smeared because different sizes of polyA tail (10–1,000 bp) were added to eGFP mRNA during polyA tailing.
Abbreviations: bp, base pairs; eGFP, enhanced green fluorescence protein; EMSA, electrophoretic mobility shift assay.
![Figure S2 Electrophoretic mobility shift assay shows complexation of mRNA and CRPPR-R9.Notes: EMSA was conducted to investigate if CRPPR-R9 complexes with mRNA. A mixture of CRPPR-R9 and mRNA produced visible retarded RNA bands from a molar charge ratio of one ([positive amines in peptide] to [phosphates in mRNA]) and a complete complexation was observed in a molar charge ratio of 4 or above. Unbound mRNA bands look smeared because different sizes of polyA tail (10–1,000 bp) were added to eGFP mRNA during polyA tailing.Abbreviations: bp, base pairs; eGFP, enhanced green fluorescence protein; EMSA, electrophoretic mobility shift assay.](/cms/asset/c842eca2-3c6e-486a-8c7c-c07436e2dcd3/dijn_a_75124_sf0002_b.jpg)
Figure S3 Gel electrophoresis after third wash step shows CRPPR-R9 incorporation in C-Lipo.
Notes: To study whether the CRPPR-R9 peptide forms a complex together with mRNA and lipofectamine, C-Lipo formation and gel electrophoresis were conducted. C-Lipo was formulated as described in the manuscript, centrifuged at 14,000× g for 30 minutes, and the supernatant containing unbound CRPPR-R9 was removed. PBS (1 mL) was added to the solution, centrifugation repeated, and the supernatant removed. This wash step was conducted twice. During the third wash step, the pellet was redispersed with 20 μL PBS, centrifuged, and the solution was carefully collected into pellet and supernatant for gel electrophoresis. The supernatant of the wash, liposome pellet, and positive control were analyzed with SDS-PAGE gel electrophoresis. The gel was stained with SYBR Green and Coomassie Blue. The third wash solution did not stain any mRNA or CRPPR, showing that the wash steps were properly conducted. Positive control of the mRNA and CRPPR peptide shows that C-Lipo contains mRNA and CRPPR peptide. Moreover, unlike the supernatant of the centrifuged C-Lipo that could not transfect cardiac fibroblasts, pelleted C-Lipo transfected cardiac fibroblasts and generated eGFP. The results prove that the CRPPR peptide is associated in C-Lipo. Scale of width of each image is 50 μm.
Abbreviations: eGFP, enhanced green fluorescence protein; PBS, phosphate buffered saline; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
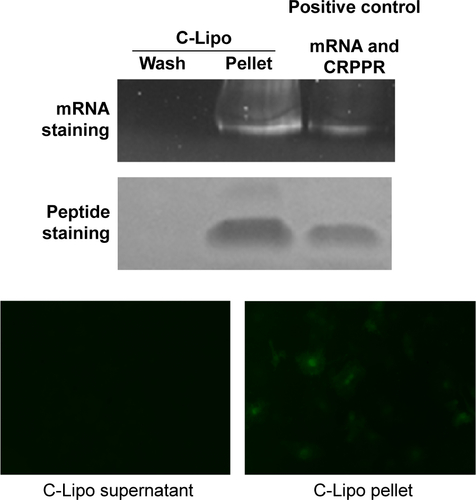
Figure S4 eGFP transfection efficiency shows that the CRPPR-R9 peptide does not improve lipofectamine transfection in HeLa cells, cervical carcinoma cells, which are not from cardiac origin.
Notes: eGFP mRNA transfection in HeLa cells was analyzed by flow cytometry. eGFP mRNA transfection efficiency of lipofectamine, CRPPR-R9/Lipo, and CRRPP-R9(scramble)/Lipo are shown. Both CRRPP-R9 and CRPPR-R9 transfection with lipofectamine show even lower transfection efficiencies compared to the lipofectamine only transfection. The results suggest that the CRPPR-R9 peptide is not effective in HeLa cells that are not targeted by the CRPPR peptide sequence. We speculate that the very low transfection efficiency of peptide/lipofectamine in HeLa cells is due to a charge imbalance that interferes with the complexation of lipofectamine and mRNAs. The results represent mean ± SE, n=3. CRPPR-R9 or CRRPP-R9 (3.8 μg, 20 molar charge ratio) was added to the eGFP mRNA (0.5 μg), and incubated at room temperature for 15 minutes. Next, 1 μL of lipofectamine 2000 in 50 μL OptiMEM was added to the solution and further incubated for 15 minutes. Cardiac fibroblasts were transfected overnight as described earlier. At 24 hours post-transfection, flow cytometry analysis was conducted. Transfection efficiency was quantified based on the percentage of eGFP+ cardiac fibroblasts using InCyte software (EMD Millipore, Billerica, MA, USA).
Abbreviations: eGFP, enhanced green fluorescence protein; SE, standard error.
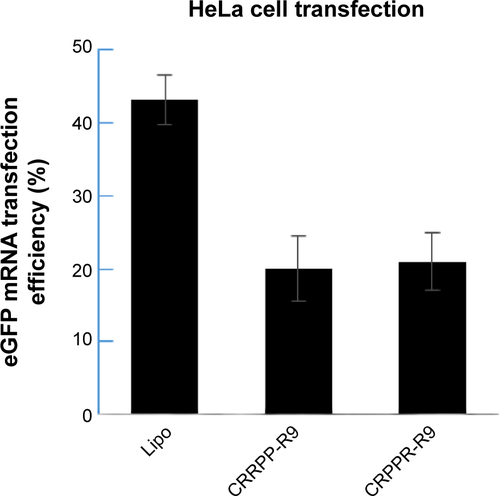
Figure S5 CRPPR-R9 does not affect cardiac fibroblast viability.
Notes: The cell viability of cardiac fibroblasts at various concentrations of CRPPR-R9 was measured with CCK-8. CRPPR-R9 and lipofectamine concentrations were determined based on transfection conditions (CRPPR 1× =3.8 μg of CRPPR-R9 per well of a 24-well plate; Lipo 5× =5 μL per well of a 24-well plate). The result shows that CRPPR-R9 does not compromise cell viability even at five-fold higher concentration than the concentration used for transfection, whereas a similar concentration level of lipofectamine caused significant cell death in 4 days.
Abbreviation: CCK-8, Cell Counting Kit-8.
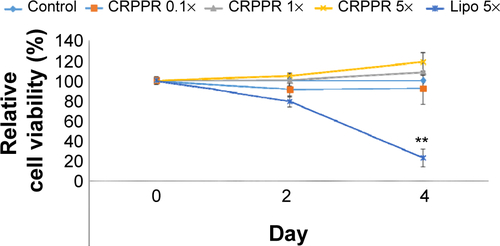
Figure S6 Low cytotoxicity of C-Lipo transfection is due to the reduced amount of lipofectamine.
Notes: Cell viability of transfected cardiac fibroblasts was measured on day 14 from the initial single transfection. Transfection was conducted with GMT mRNAs as described in the main manuscript. One-quarter of the Lipo sample contained one-quarter of the lipofectamine amount used for Lipo. The C-Lipo and 1/4 Lipo transfections showed similar levels of cell viability. This result suggests that the high cell viability of the C-Lipo transfection is mainly due to the reduced amount of lipofectamine.
Abbreviation: GMT, Gata4, Mef2c, and Tbx5.
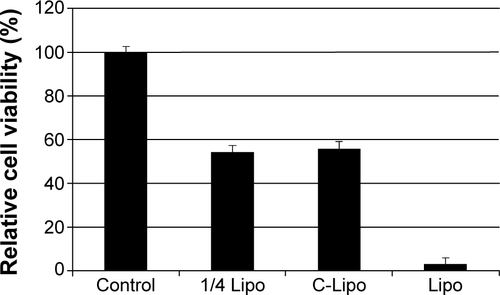
Table S1 Primer list
