Figures & data
Figure 1 Schematic illustration of self-protection by mammalian cells during cationic polymers-mediated transfection.
Abbreviations: CM, cell membrane; DNA, deoxyribonucleic acid.
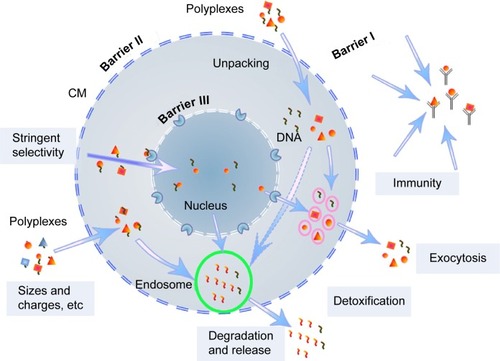
Table 1 Weight ratio of the reactants and molecular weights of PG6-PEI-INO polymers
Figure 2 Synthesis route of PG6-PEI-INO polymers.
Abbreviations: CMPG6, carboxymethyl polyglycerol; CMINO, carboxymethyl inositol; DCC, N,N’-dicyclohexylcarbodiimide; DMF, N-dimethyl-formamide; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydro chloride; INO, myo-inositol; NHS, N-hydroxysuccinimide; PEI, polyethylenimine; PG6, polyglycerol.
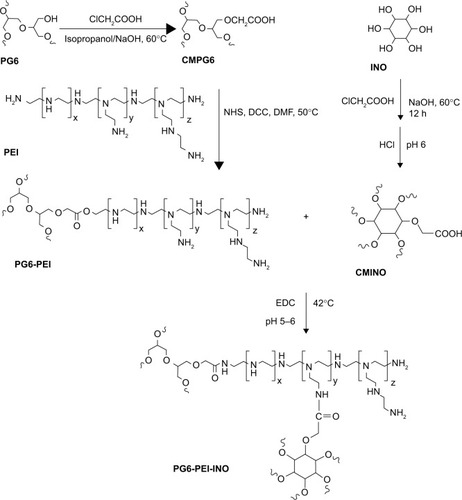
Figure 3 Characterization of the PG6-PEI-INO polymers.
Notes: (A) FT-IR spectra of PG6-PEI-INO 1, PG6-PEI-INO 2, and PG6-PEI-INO 3. (B) 1H NMR spectra of the polymers. Deuterium oxide was used as the solvent.
Abbreviations: CMINO, carboxymethyl inositol; CMPG6, carboxymethyl polyglycerol; FT-IR, Fourier transform infrared spectroscopy; INO, myo-inositol; NMR, nuclear magnetic resonance; PEI, polyethylenimine; PG6, polyglycerol.
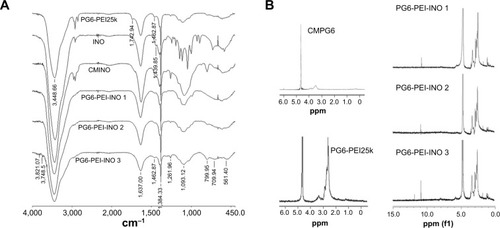
Figure 4 DNA-binding ability of PG6-PEI-INO polymers.
Notes: (A) Agarose gel electrophoresis of PG6-PEI-INOs/pEGFP-C1 complexes at varied weight ratios. (B) Morphologic study of PG6-PEI-INO/pEGFP-C1 (w/w =5) complexes using transmission electron microscopy.
Abbreviations: INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol.
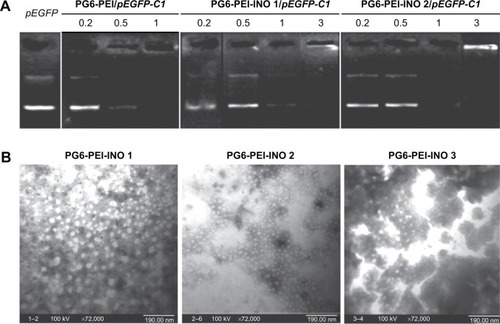
Figure 5 Biocompatibility of the PG6-PEI-INO/plasmid or PG6-PEI-INO polymers.
Notes: (A) MTT analysis of cell viability was performed after 293T cells were cocultivated for 52 hours with PG6-PEI-INOs/plasmid (pEGFP-C1), which had weight ratios of 5 (a), 7 (b), and 9 (c), respectively. (B) MTT analysis of cell viability was performed after 293T cells were cocultivated with identical dosages of PG6-PEI-INOs for 52 hours. Cells were treated with PEI25k/pEGFP-C1 at a weight ratio of 6, or with an identical dosage of PEI25k as the control. DNA was used at 1.3 μg of pEGFP-C1 per mL medium.
Abbreviations: INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol; p, plasmid (pEGFP-C1).
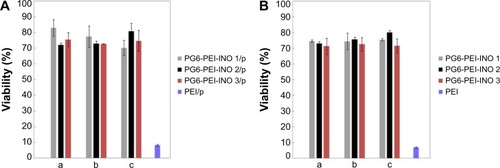
Figure 6 Transfection activities of PG6-PEI-INO polymers.
Notes: (A) Transgene expression reported by protein EGFP in 293T cell line treated with (a) PG6-PEI-INO 1/pEGFP-C1, (b) PG6-PEI-INO 2/pEGFP-C1, and (c) PG6-PEI-INO 3/pEGFP-C1 complexes at weight ratio of 7, respectively. Plasmid pEGFP-C1 was used at 1.3 μg per mL culture medium. The cells were cultivated for 52 hours. The adherent cells and the cell suspensions achieved with trypsin treatment were analyzed by phase-contrast fluorescent microscopy. Scale bar: 15 μm. (B) EGFP-positive cell ratios achieved by transgene expression mediated by PG6-PEI-INO polymers. The 293T cells were transfected using the following agents: (a) PG6-PEI-INO 1, (b) PG6-PEI-INO 2, (c) PG6-PEI-INO 3, (d) PG6-PEI at varied weight ratios to pEGFP-C1, and (e) the PEI25k control at its optimal weight ratio (1.3) to pEGFP-C1 (1.3 μg of pEGFP-C1 per mL cell culture).
Abbreviations: INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol; EGFP, enhanced green fluorescent protein.
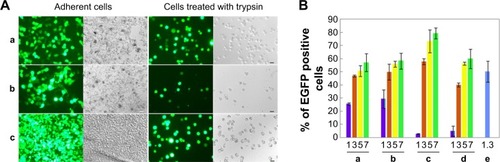
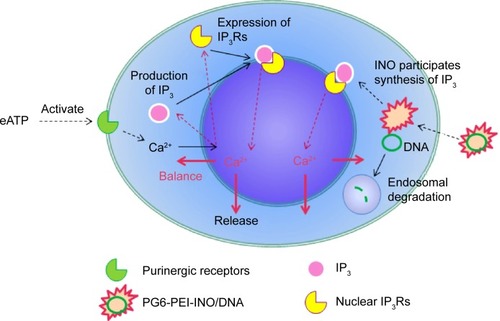
Figure 7 Effect of extracellular ATP on the growth rate of 293T cells transfected with various polymer/complexes (at optimal weight ratio for transfection).
Notes: MTT analyses of cell viability were performed after the 293T cells were cocultivated with the materials for 44 hours. About 1.3 μg of plasmid pEGFP-C1 was used per microliter of cell culture.
Abbreviations: ATP, adenosine triphosphate; INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol.
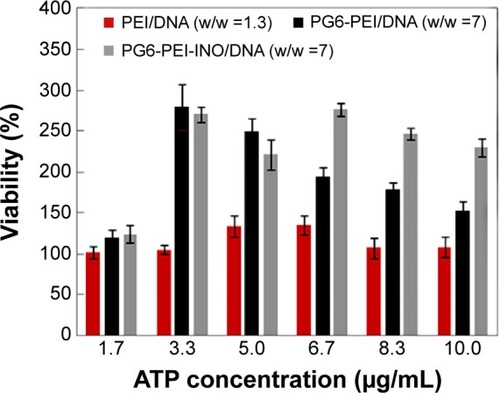
Figure 8 Inhibition of extracellular ATP on transgene activity of PG6-PEI-INO.
Notes: (A) The 293T cells were treated with (a) PEI25k/pEGFP-C1 (w/w =1.3), (b) PG6-PEI/pEGFP-C1 (w/w =7), and (c) PG6-PEI-INO 3/pEGFP-C1 (w/w =7) for 96 hours, respectively. Extracellular ATP was used at 1.7 μg per mL culture medium. Scale bar: 10 μm. (B) EGFP-positive 293T cell ratios mediated by polymers/DNA/ATP. Plasmid DNA (pEGFP-C1) was used at 1.3 μg per mL culture medium. The cells were treated with the polymers/DNA/ATP for 44 hours (a) and 96 hours (b), respectively.
Abbreviations: ATP, adenosine triphosphate; INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol.
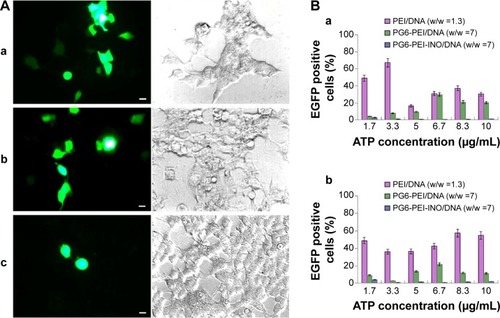
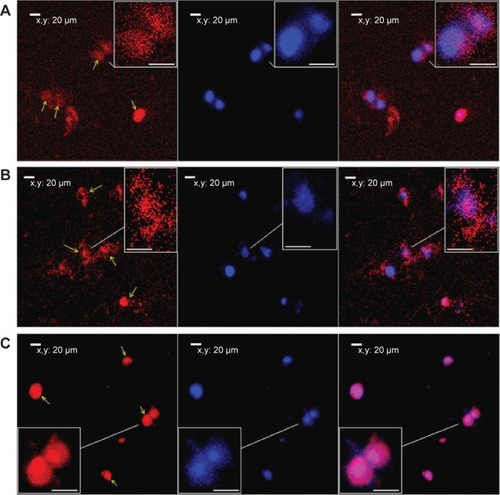
Figure 9 Fluorescence tracking of PG6-PEI-INO polymers in 293T cell nuclei.
Notes: Fluorescence was analyzed after the 293T cells were cocultivated for 48 hours with (A) PG6-PEI-INO-Rh 1, (B) PG6-PEI-INO-Rh 2, and (C) PG6-PEI-INO-Rh 3, which had increased inositol ligands per molecule (1:1, 1:10, and 1:35, respectively). Accumulation of PG6-PEI-INO-Rhs in cell nuclei was indicated with arrows. Scale bar in magnified images: 15 μm. Red fluorescence shows Rhodamine B (Emission: ~572 nm) labeled polymers. Blue fluorescence shows the cell nuclei stained with DAPI (Emission: ~461 nm when bound with DNA). Merged images in purple color show the PG6-PEI-INO-Rh polymers in cell nuclei.
Abbreviations: DAPI, (4′,6-diamidino-2-phenylindole); INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol; Rh, Rhodamine.