Figures & data
Figure 1 Production of PDMS molds and casting of nanostructured PLGA films: (A) 190 nm, 300 nm, 400 nm, and 530 nm PS beads were dispersion deposited on glass coverslips; (B) Coverslip fixed to equal diameter glass rod (green); (C) Coverslip/glass rod fixed to Petri dish; (D) PDMS (orange) poured to fill Petri dish; (E) PDMS from the previous step removed from the Petri dish, inverted, and placed in a larger Petri dish (red), with additional PDMS (orange), leaving a well in PDMS (white) with a nanotextured bottom; (F) PLGA in chloroform (blue) poured into well then allowed to evaporate; and (G) Dried PLGA removed from the well and inverted, leaving PLGA surfaces with spherical nanometer surface features. Copyright © 2008 obtained with permission from Carpenter J, Khang D, Webster TJ. Nanometer polymer surface features: The influence on surface energy, protein adsorption and endothelial cell adhesion. Nanotechnology. 2008;19(50):505103.
Abbreviations: PDMS, polydimethylsiloxane; PLGA, poly-lactic-co-glycolic acid; PS, polystyrene.
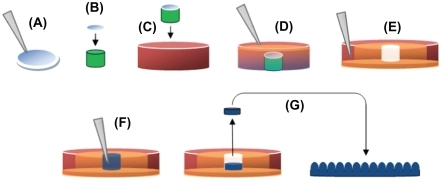
Figure 2 AFM images and RMS values: (A–B) AFM images of PLGA surfaces created by solution evaporation with (A) 0.5 g PLGA:8 mL chloroform, (B) 0.5 g PLGA:4 mL chloroform. (C–F) AFM images of PLGA surfaces created by using templates of PS nanobeads with a diameter of (C) 190 nm, (D) 300 nm, (E) 400 nm and (F) 530 nm.
Abbreviations: AFM, atomic force microscopy; RMS, root mean square roughness; PLGA, poly-lactic-co-glycolic acid; PS, polystyrene.
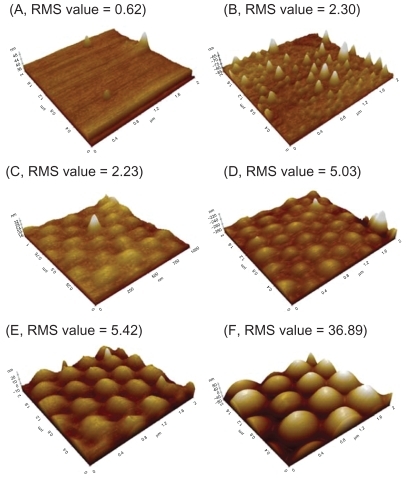
Figure 3 dH2O static contact angles on PLGA samples. Static contact angles were obtained immediately after the deposition of a 3 μL drop of dH2O under ambient conditions.
Notes: Values are mean ± SEM, n = 3. *P < 0.01, **P < 0.05 and ***P < 0.1.
Abbreviations: dH2O, distilled water; PLGA, poly(lactic-co-glycolic) acid; SEM, standard error of the mean.
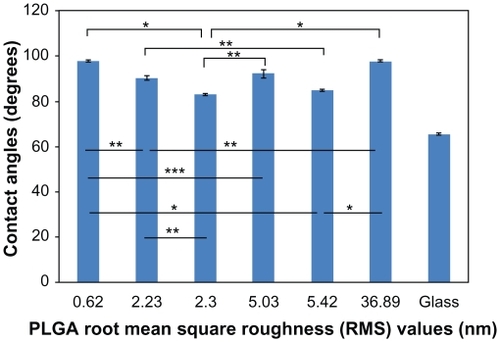
Figure 4 Lung cancer epithelial cell adhesion results on the various PLGA samples and borosilicate glass. Data expressed as mean ± standard deviation of the mean.
Notes: *P < 0.01, **P < 0.05 and **P < 0.1. Less lung carcinoma cells adhered to the PLGA surfaces with an RMS value of 5.42 nm, compared with the surfaces with an RMS value of 2.30 nm (P < 0.05). Comparing the PLGA surfaces with RMS values of 2.30, 2.23, and 5.42 nm, the least cells adhered to the surfaces with an RMS value of 5.42 nm. PLGA surfaces with RMS values of 5.42, 0.62, and 2.23 nm had the least cancer cell adhesion.
Abbreviations: PLGA, poly(lactic-co-glycolic) acid; RMS, root mean square roughness.
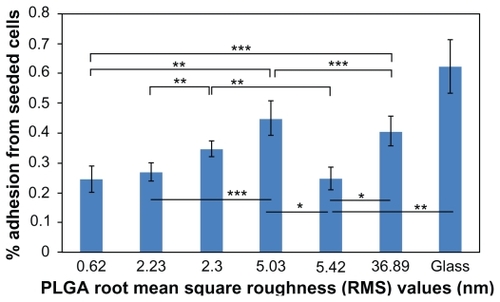
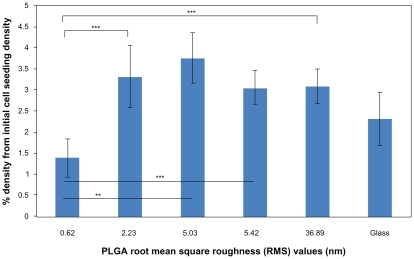
Figure 5 Lung cancer epithelial cell proliferation results on the various PLGA samples and borosilicate glass. Cell seeding density was 50,000 cells/cm2.
Notes: Data expressed as mean ± standard deviation of the mean. **P < 0.05 and ***P < 0.1. The PLGA surfaces with an RMS value of 0.62 nm had lower cancer cell densities after three days than any other nanorough PLGA surfaces.
Abbreviations: PLGA, poly(lactic-co-glycolic) acid; RMS, root mean square roughness.