Figures & data
Figure 2 X-Tile analysis of survival data from the SEER database. (A) The X-Tile plot displays red coloration of cut-points, indicating a negative correlation with survival, while green coloration represents a positive correlation. The plot illustrates patient age divided at the mean age, and the optimal cut-point is highlighted by a black circle located in the middle of the color bar. (B) The histogram of the entire cohort reveals that the optimal cut-off value of age at diagnosis is 73 years, based on cancer-specific survival (CSS). The X-axis represents the age of patients, and the Y-axis represents the number of patients at each age. (C) The Kaplan-Meier survival curve is generated based on these cut-off values.
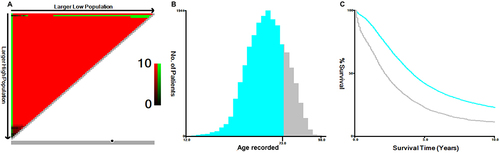
Table 1 Baseline Characteristics of Patients with Advanced Ovarian Serous Cystadenocarcinoma in the SEER Database, n(%)
Table 2 The Cox Hazard Regression Analysis of Overall Survival
Table 3 The Cox Hazard Regression Analysis of Cancer-Specific Survival
Figure 3 Kaplan-Meier curves of overall survival (OS) and cancer-specific survival (CSS). Kaplan-Meier curves of OS (A) and CSS (B) before propensity score matching (PSM), Kaplan-Meier curves of OS (C) and CSS (D) after PSM.
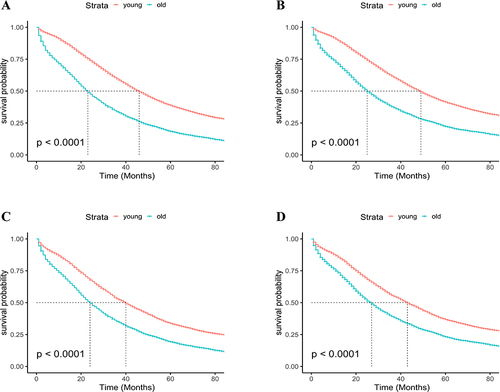
Figure 4 The forest plot of subgroup analyses according to age (young vs old) of patients with advanced serous ovarian cystadenocarcinoma in the PSM cohort from the SEER database.
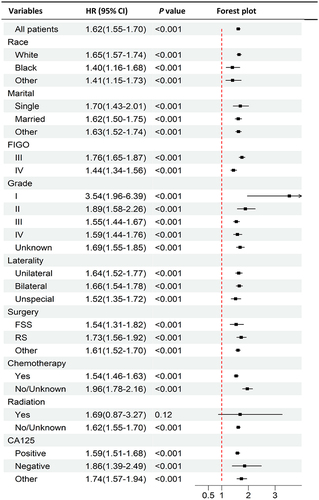
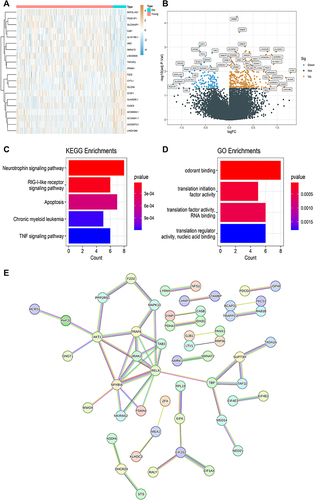
Figure 5 Differentially expressed genes (DEGs) between the old group and the young group. (A) The heatmap of DEGs. (B) The volcano plot of DEGs. (C) A bar chart of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of DEGs. (D) A bar chart of Gene Ontology (GO) analysis of DEGs. (E) The protein-protein interaction network of DEGs.
Data Sharing Statement
The following information was supplied regarding data availability: Data is available at the SEER database (https://seer.cancer.gov/) and the TCGA database (https://portal.gdc.cancer.gov/).