Figures & data
Table 1 Study population
Figure 1 Self-reported age of asthma onset.
Abbreviation: SNP, single-nucleotide polymorphism.
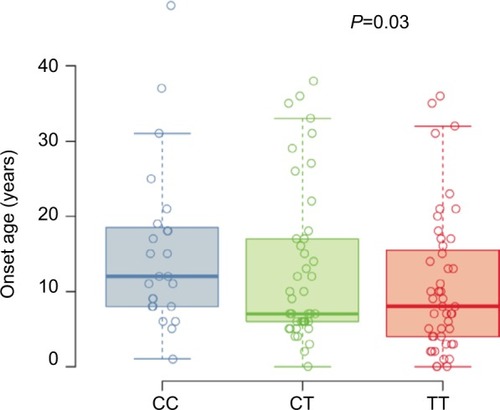
Figure 2 FeNO.
Abbreviations: FeNO, fractional exhaled nitric oxide; SNP, single-nucleotide polymorphism.
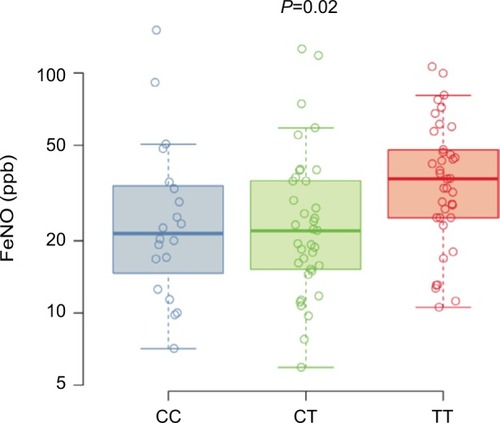
Figure 3 Eosinophil counts.
Abbreviation: SNP, single-nucleotide polymorphism.
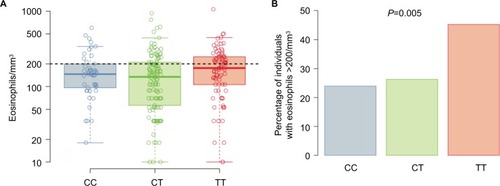
Figure 4 ORMDL3 expression.
Abbreviations: EOS, eosinophils; PBMCs, peripheral blood mononuclear cells.
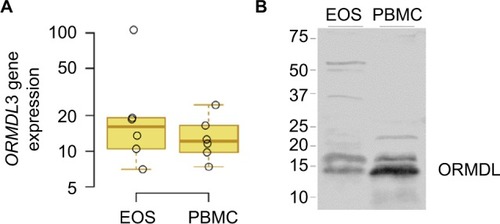
Figure S1 FeNO.
Notes: Box plots for the FeNO measured in ppb for the Caucasian non-Hispanic subgroup (n=120) stratified by genotype at SNP rs7216389, CC, CT, and TT. Boxes denote the median and 25th to 75th percentiles with whiskers extending to the farthest points within 1.5 times the box height. The P-value indicates the significance of the association between the number of T alleles and FeNO levels.
Abbreviations: FeNO, fractional exhaled nitric oxide; SNP, single-nucleotide polymorphism.
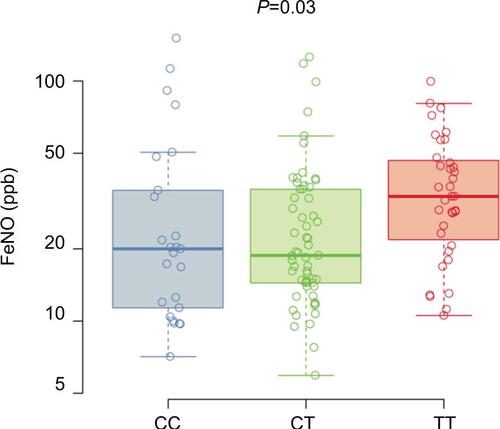
Figure S2 Percentage of donors in Caucasian non-Hispanic cohort (n=176) with geometric mean eosinophil counts >200/mm3 stratified genotype at SNP rs7216389, CC, CT, and TT. The P-value indicates the significance of the association between the number of T alleles and the percentage of individuals with eosinophil counts >200/mm3.
Abbreviation: SNP, single-nucleotide polymorphism.
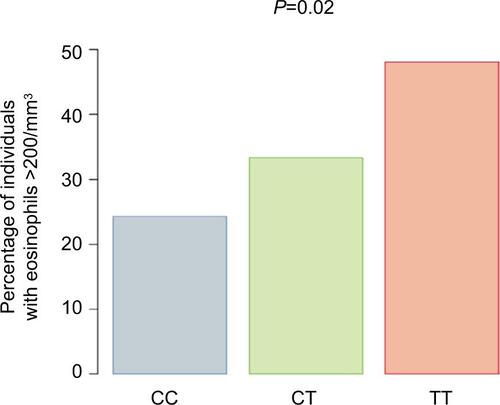
Table S1 FeNO study population
