Figures & data
Figure 1 Expression profiles of PIGC mRNA in human malignant tumors. (A) Oncomine database, (B) GEPIA website.
Figure 2 Correlation between PIGC mRNA expression and PIGC DNA methylation. (A) Heat map of PIGC expression and its DNA methylation in cancerous liver specimens and normal liver tissues. (B) PIGC mRNA expression is higher in cancerous liver tissues (N=372) than that in normal liver tissues (N=49).
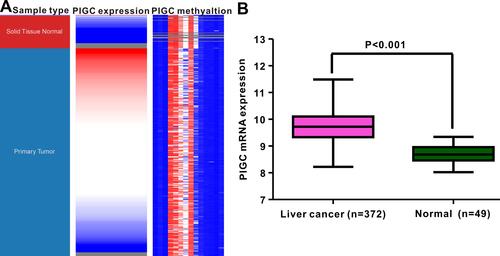
Figure 3 Association between expression of PIGC mRNA and clinical characteristics in liver cancer from the UALCAN database. (A) Gender, (B) age, (C) tumor grade, (D) N stage, (E) TNM stage, (F) TP53 mutation.
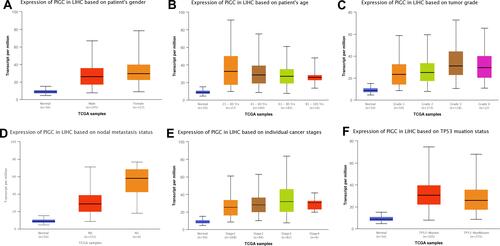
Figure 4 Association between PIGC expression and survival in patients with liver cancer. Overexpression of PIGC correlates with worse overall survival (A) and disease-free survival (B) in patients with liver cancer. PIGC overexpression is correlated with worse overall survival in patients without viral hepatitis (C), but not in patients with viral hepatitis (D). Overexpression of PIGC is correlated with worse disease-free survival in patients without viral hepatitis (E), but not in patients with viral hepatitis (F).
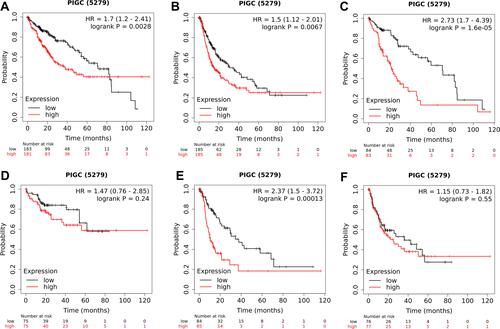
Figure 5 Relationship between PIGC methylation and PIGC mRNA expression in liver cancer. (A) Levels of PIGC DNA methylation are lower in cancerous liver tissues (N=379) compared with that in normal liver tissues (N=50). (B) A negative correlation (r=−0.398, P<0.0001) is observed between PIGC DNA methylation and PIGC mRNA expression. (C) Distribution of 14 PIGC DNA CpG sites in liver cancer and normal tissues.
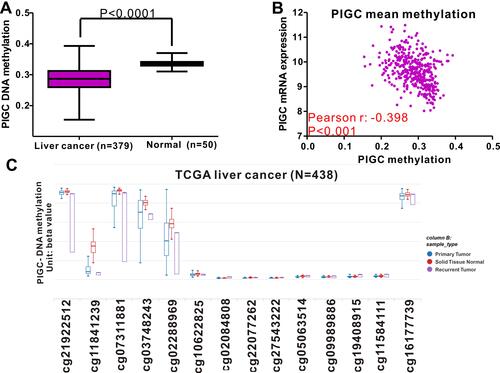
Figure 6 PIGC gene mutation rate and its clinical relevance in liver cancer. (A) The mutation rate of PIGC is 10%. PIGC mutation is significantly correlated with T stage (B), M stage (C), surgical margin resection status (D), and fraction genome alteration (E).
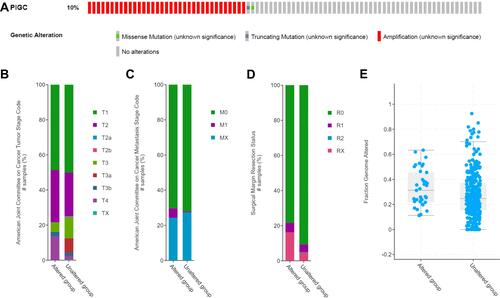
Figure 7 Expression of PIGC and effects of PIGC on proliferation and migration in liver cancer cell lines. (A) Expression of PIGC protein is higher in liver cancer cell lines than in normal liver cell line. (B) Si-PIGC-2 is the most efficient Si-RNA revealed by Western blot. (C) PIGC contributes to the migration of cancerous liver cells. (D) Quantitative analysis of migrated cells in four groups. (E) PIGC contributes to the proliferation of cancerous liver cells. *Stands for the P value less than 0.05.
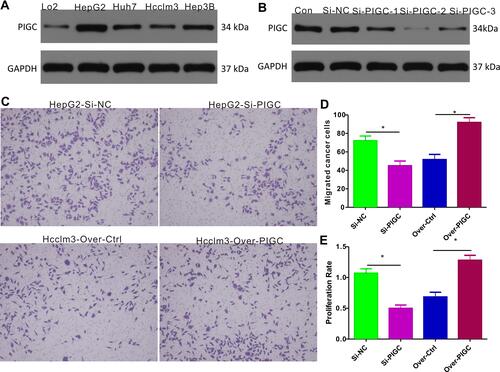
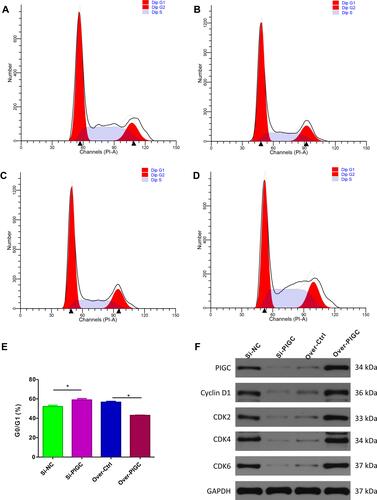
Figure 8 Effects of PIGC on the cell cycle in liver cancer cell line. (A–D) Flow cytometric analysis showed that G0/GI ratio was increased in PIGC-silenced HepG2 cell line (B) compared to the Si-NC cell line (A). G0/GI ratio was dramatically reduced in PIGC-overexpressed Hcclm3 cell line (D) compared to the Hcclm3 cell line transfected with the empty vector (C). Quantitative analysis of G0/GI ratio in four groups (E). Expression of CDK protein in PIGC-silenced and PIGC-overexpressed cell lines (F). *Stands for the P value less than 0.05.