Figures & data
Table 1 Demographics of normal human subjects (n = 12)
Table 2 Real-time PCR primers
Figure 1 Whole blood was stimulated with 100 nM dexamethasone (A) or 1 ng/mL LPS with and without 100 nM dexamethasone (B) for 2 hours. RNA was isolated and cDNA transcribed. Real-time PCR was performed to determine expression of the GR upregulated genes GILZ, FKBP51, FLAP, and MKP-1 (A) and the GR downregulated genes IL-6, TNFα, and IFNγ (B). The expression of these genes was normalized to the housekeeping genes GAPDH and CAP-1. ΔΔ Ct was calculated for GILZ, FKBP51, FLAP, and MKP-1 as treatment (dexamethasone) against no treatment (A) and for IL-6, TNFα, and IFNγ as treatment (dexamethasone + LPS) against LPS only.
Abbreviations: LPS, lipopolysaccharide; PCR, polymerase chain reaction; GR, glucocorticoid receptor; GILZ, glucocorticoid inducible leucine zipper; FKBP51, FK506 binding protein 51; FLAP, arachidonate 5-lipoxygenase-activating protein; MKP- 1, mitogen activated protein kinase phosphatase 1; IL-6, interleukin 6; TNFα, tumor necrosis factor α; IFNγ, interferon γ; IQR, interquartile range; CAP-1, cyclic AMPaccessory protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
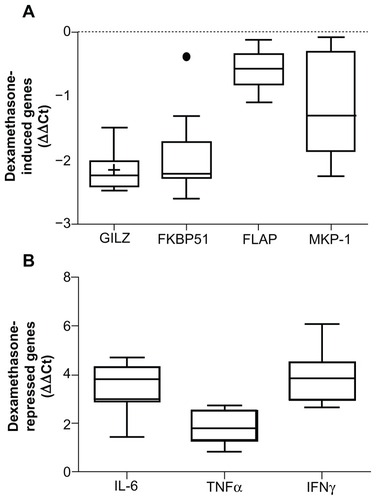
Figure 2 Whole blood was stimulated with increasing amounts of dexamethasone (A) or 1 ng/mL LPS with and without increasing concentrations of dexamethasone (B) for 2 hours. RNA was isolated and cDNA transcribed. Real-time PCR was performed to determine expression of the GR upregulated genes GILZ and FKBP51 (A) and the GR downregulated genes IL-6 and TNFα (B). The expression of these genes was normalized to the housekeeping genes GAPDH and CAP-1. Dose response curves were drawn and the EC50 (A) and IC50 (B) values were calculated.
Abbreviations: LPS, lipopolysaccharide; PCR, polymerase chain reaction; GR, glucocorticoid receptor; GILZ, glucocorticoid inducible leucine zipper; IL-6, interleukin 6; TNFα, tumor necrosis factor α; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CAP-1, cyclic AMP-accessory protein; EC50, half maximal effective concentration; IC50, half maximal inhibitory concentration; IQR, interquartile range.
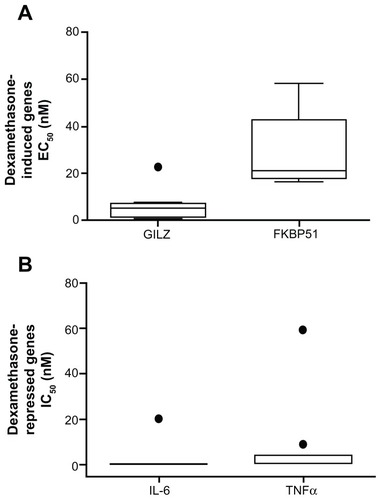
Figure 3 Whole blood was drawn from 10 individuals at 8:30 am, 12:30 pm, 4:30 pm, and 8:30 pm and was stimulated with 100 nM dexamethasone (A and B) or 1 ng/mL LPS with and without 100 nM dexamethasone (C and D) for 2 hours. RNA was isolated and cDNA transcribed. Real-time PCR was performed to determine expression of the GR upregulated genes GILZ (A) and FKBP51 (B) and the GR downregulated genes IL-6 (C) and TNFα (D).
Abbreviations: LPS, lipopolysaccharide; PCR, polymerase chain reaction; GR, glucocorticoid receptor; GILZ, glucocorticoid inducible leucine zipper; FKBP51, FK506 binding protein 51; IL-6, interleukin 6; TNFα, tumor necrosis factor α; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CAP-1, cyclic AMP-accessory protein; ΔΔCt, comparative cycle threshold.
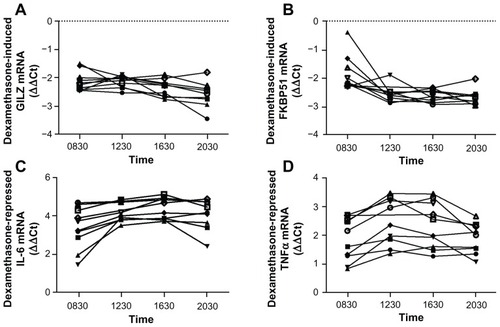
Figure 4 Whole blood from 10 individuals was either treated immediately (closed square) or stored in the EDTA tube at 4°C for 24 hours (open square) prior to stimulation with 100 nM dexamethasone (A) or 1 ng/mL LPS with and without 100 nM dexamethasone (B) for 2 hours. RNA was isolated and cDNA transcribed. Real-time PCR was performed to determine the expression of the GR upregulated genes GILZ and FKBP51 (A) and the GR downregulated genes IL-6 and TNFα (B).
Abbreviations: LPS, lipopolysaccharide; PCR, polymerase chain reaction; GR, glucocorticoid receptor; GILZ, glucocorticoid inducible leucine zipper; FKBP51, FK506 binding protein 51; IL-6, interleukin 6; TNFα, tumor necrosis factor α; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CAP-1, cyclic AMP-accessory protein; ΔΔCt, comparative cycle threshold.
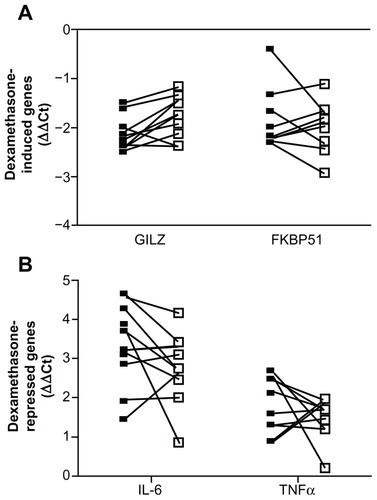
Figure 5 Whole blood was drawn at 8:30 am and stimulated with 1 ng/mL LPS with and without 100 nM dexamethasone for 2 hours for real-time PCR or 24 hours for ELISA.
Abbreviations: LPS, lipopolysaccharide; PCR, polymerase chain reaction; EL ISA, enzyme-linked immunosorbent assay; IL-6, interleukin 6; GAPDH, glyceraldehyde- 3-phosphate dehydrogenase; CAP-1, cyclic AMP-accessory protein; SD, standard deviation.
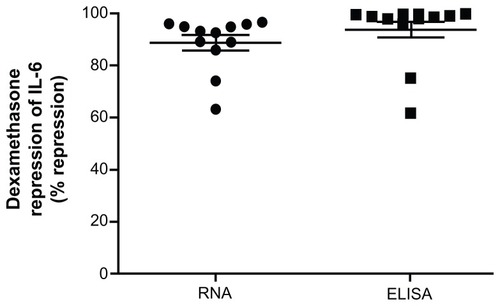
Table 3 Coefficient of variation over gene
Table 4 Descriptive statistics by figure, gene, and time