Figures & data
Table 1 Physiological Parameters of Experimental Mice Groups
Table 2 Biological Parameters of Experimental Mice Groups
Figure 1 MLT reversed HFD-mediated hepatic anomalies. Male C57BL/6 mice were administered high fat diet (HFD) or a normal control diet (NCD) for 6 months, together with MLT (10mg/kg/day, 20mg/kg/day) for every alternative day. (A) Relative body weight changes in different groups. (B) Representative images of Liver of different groups, (C and D) cross-sections of Liver with HE and MT staining in the different groups. Scale bars = 100 μm. (E–I) Histology score of different groups, The values indicate the mean ± SEM (n=3) ***P<0.001, ****P<0.0001.
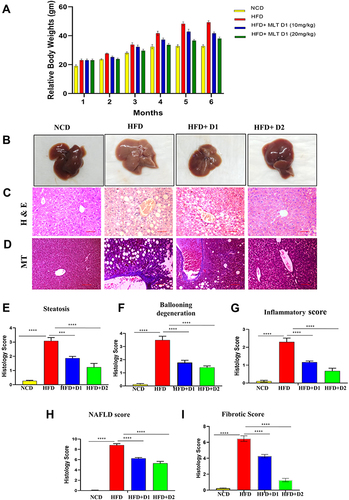
Figure 2 MLT reduced HFD-induced Kupffer cell activation. (A) Characterization of Kupffer cells by flow cytometric analysis using F480-FITC and CD11b-PE dual staining (B) Change of the activated Kupffer cells population in HFD and MLT treated groups and simultaneously Pro-IL 1β level also checked by flow cytometry using Pro-IL-1β-APC. (C) Bar graph of Pro-IL1β population observed from Kupffer cells (D) Kupffer cell were isolated from the mice liver and IL-1β was measured by ELISA from the cell supernatant. The values indicate the mean ± SEM (n=3). *p < 0.05, ****P<0.0001.
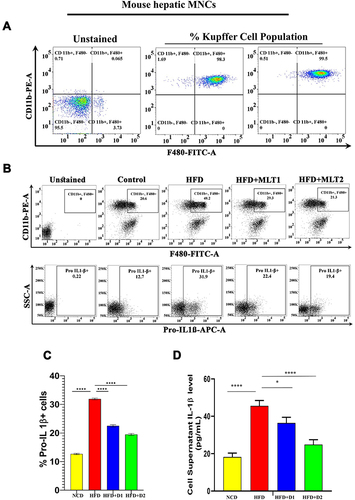
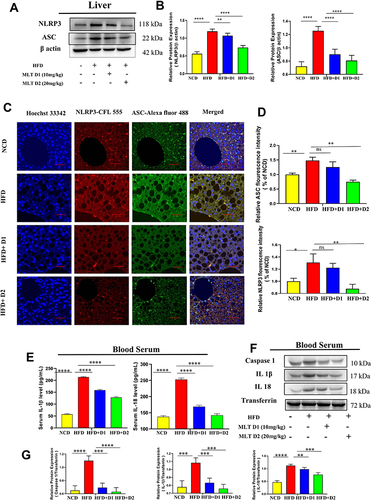
Figure 3 MLT impairs HFD-mediated inflammasome activation. (A) Western blot analysis of inflammasome assembly protein NLRP3 and ASC from the liver of different mice groups. (B) The bar graph showed the quantification of NLRP3/β actin and ASC /β actin, respectively. (C) Representative immunofluorescent images showed NLRP3 and ASC expression in mouse liver depending on the different treatments. Scale bar = 50 μm. (D) The bar graph represents quantification of the fluorescence intensity of NLRP3 and ASC shown in the different groups. (E) level of IL-1β and IL-18 was analyzed by ELISA from different mice groups. (F) Western blot analysis of pro-inflammatory cytokines (Caspase 1, IL-1β, and IL-18) in the serum of different groups. (G) The bar graph showed the quantification of Caspase 1/Transferrin, IL-1β /Transferrin, and IL-18/Transferrin, respectively. (H) ELISA of IL-1β and IL-18 in cell supernatant of OA/PA administered HepG2 cells, with or without MLT (I) Western blot analysis of NLRP3 and ASC in the OA/PA administered HepG2 cells, with or without MLT. (J) The bar graph showed the quantification of NLRP3/β actin and ASC /β, respectively in FFA treated HepG2 cell lysates. (K) Glyburide was treated with OA/PA in a different set of the experiment as a positive control and NLRP3 protein expression is shown by Western blot analysis. (L) The bar graph represents the relative protein expression of NLRP3/β actin. The values indicate the mean ± SEM (n=3), *p < 0.05, **P< 0.01, ***P<0.001, ****P<0.0001.
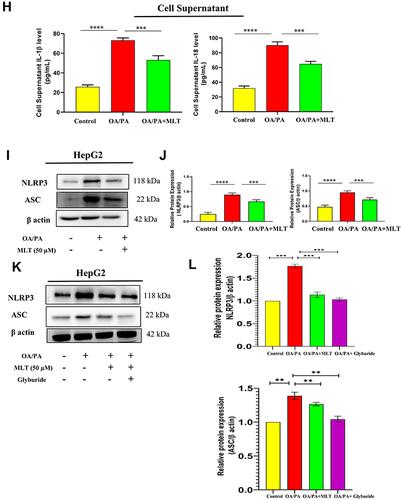
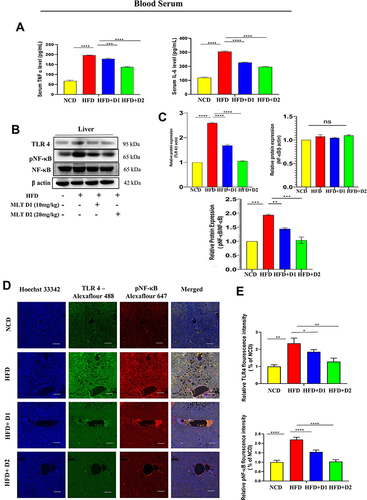
Figure 4 MLT inhibited HFD-induced TLR4-guided NF-κB activation. (A) Pro-inflammatory cytokines, such as TNF-α and IL-6 were analyzed from serum by ELISA from different mice groups. (B) Western blot analysis of inflammatory proteins (TLR 4, pNF-κB) in the liver of different groups. (C) The bar graph showed the quantification of TLR 4/β actin and pNF-κB/NF-κB, respectively. (D) Representative immunofluorescent images showed TLR 4, pNF-κB expression in mouse liver depending on the different treatments. Scale bar = 50 μm. (E) The bar graph represents the quantification of the fluorescence intensity of TLR 4 and pNF-κBshown in the different groups. (F) ELISA of TNF-α and IL-6 in cell supernatant of OA/PA administered HepG2 cells, with or without MLT. (G) Western blot analysis of inflammatory proteins (TLR 4, pNF-κB) in the HepG2 cell lysate of different groups. (H) The bar graph exhibits relative protein expression of TLR 4/β actin and p-NF-κB/ NF-κB. The values indicate the mean ± SEM (n=3), *p < 0.05, **P< 0.01, ***P<0.001, ****P<0.0001.
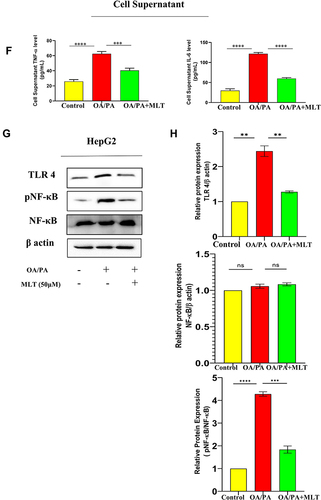
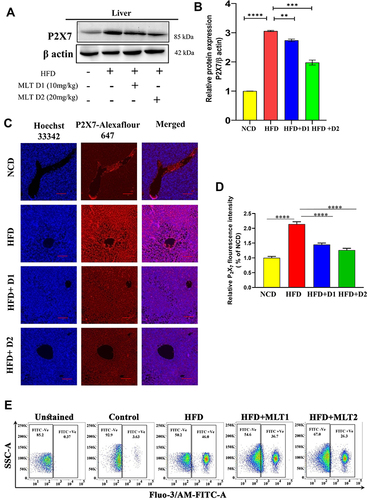
Figure 5 MLT suppressed HFD-induced P2X7 receptor activation and intercellular Ca2+ level. (A) Western blot analysis of P2X7 protein from the liver of different mice groups, (B) The bar graph represents the relative protein expression of P2X7/β actin. (C) The immunofluorescent image represents the P2X7 protein expression in the liver of different mice groups, Scale bar = 50 μm. (D) The bar graph shows the relative fluorescence intensity of P2X7. (E) This data represents intracellular Ca2+ ions in the primary liver cells of different mice groups. (F) Western blot analysis of P2X7 protein from HepG2 cell lysates administered with OA/PA, with or without MLT. (G) The bar graph represents the relative protein expression of P2X7/β actin. (H) The data represents in-vitro intracellular Ca2+ ion levels. (I) the HepG2 cells were treated with known P2X7 inhibitor brilliant blue G (BBG) along with OA/OA and MLT, the Western blot analysis shows P2X7 and NLRP3 protein expression. (J) The bar graph exhibits relative protein expression of P2X7/β actin and NLRP3/β actin. (K) The immunofluorescent image depicts P2X7 protein expression in different groups (Control, OA//PA, OA/PA+MLT, OA/PA+BBG), Scale bar = 20 μm. (L) The bar graph represents the relative expression of P2X7. The values indicate the mean ± SEM (n=3), **P< 0.01, ***P<0.001, ****P<0.0001.
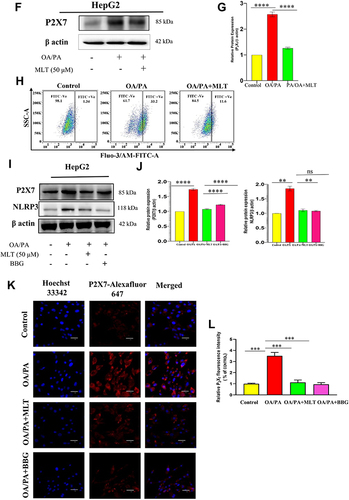
Figure 6 MLT inhibited HFD-induced iROS generation and suppressed NOX activation. (A and B) Flow cytometric analysis of intracellular ROS and bar graph of mean fluorescence unit of iROS generation, (C) Western blot analysis of NOX-2 and Nrf2 protein, and (D) the bar graph exhibits relative protein expression of NOX 2/ and Nrf2/ β actin. The values indicate the mean ± SEM (n=3). ***P<0.001, ****P<0.0001.
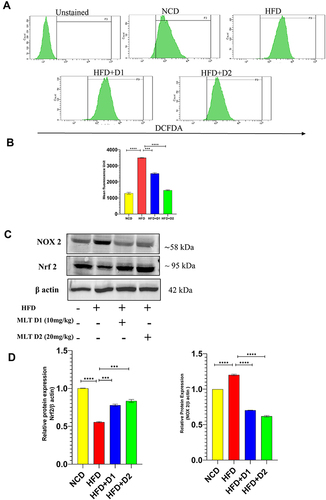
Figure 7 MLT interacted with the P2X7 receptor. (A and B) 2D and 3D image of molecular interaction study of MLT and P2X7.
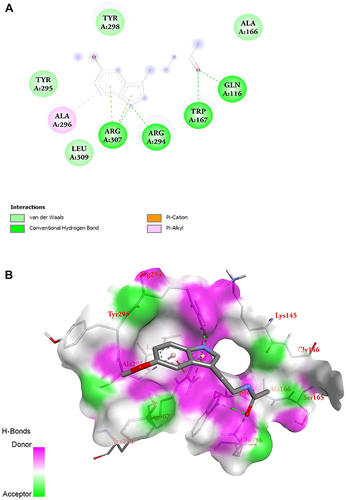
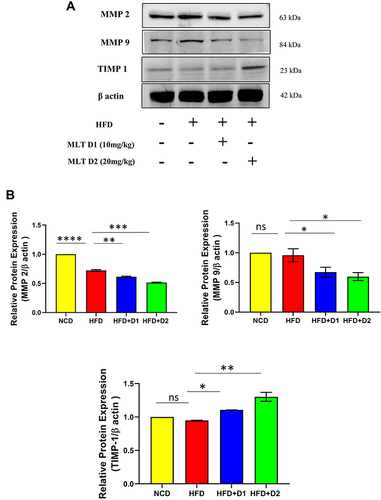
Figure 8 MLT inhibited HFD-mediated hepatic fibrosis through MMP regulation. (A) Western blot analysis of MMP2, MMP9, and TIMP 1 protein, (B) The bar graph showed the quantification of MMP 2/β actin, MMP 9/β actin, and TIMP 1/β actin, respectively. The values indicate the mean ± SEM (n=3), *p < 0.05, **P< 0.01, ***P<0.001, ****P<0.0001.