Figures & data
Figure 1 Base excision repair proteins are upregulated in PBMC from hospitalized Covid-19 patients. (A) Levels of 8-oxoG in DNA extracted from PBMC obtained from hospitalized Covid-19 patients (n = 22) and healthy controls (n = 11). 8-oxoG was measured by liquid chromatography with tandem mass spectrometry (LC-MS/MS). (B) Relationship between 8-oxoG levels and the degree of respiratory failure as assessed by pO2/FiO2-(P/F-ratio) where low P/F ratio characterize those with the most severe degree of respiratory failure. Correlation analysis by Pearson correlation coefficient. (C) Comparison of relative BER protein abundances measured by targeted MS. The error bar shows the standard deviation of measured peak area in samples from each group (control or patient). Endogenous β-actin levels were used for data normalization. A list of peptides used for quantification is provided in Supplemental File 1. Student’s t test, *p < 0.05, **p < 0.001. Values are shown as mean ± SEM. PBMC; peripheral blood mononuclear cells, BER; base excision repair. Illustration created with BioRender.com. Related to Figure S2.
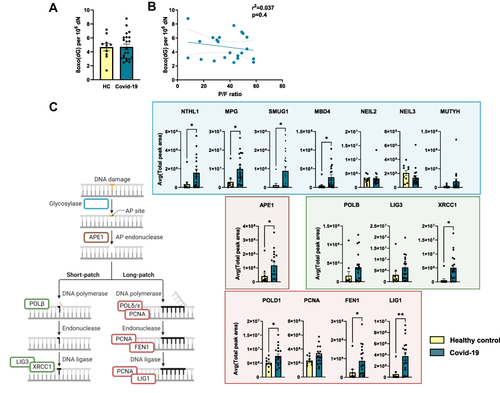
Figure 2 Enrichment analyses confirm regulation of DNA repair in PBMC from hospitalized Covid-19 patients. (A) Top 15 regulated groups of Gene Ontology (GO) terms and Reactome (R-HSA) pathways found by enrichment analysis including regulated protein coding transcripts in PBMC from Covid-19 patients (n = 15) vs healthy controls (n = 5). (B) Illustration of cellular processes affected by DNA damage response proteins linked to selected significantly regulated GO terms and Reactome pathways found in the enrichment analysis (all terms and pathways listed in. Supplemental File 2). (C) Heatmap showing the protein coding transcripts enriched in the GO term DNA repair (GO:0006281) and Reactome pathway DNA repair (R-HSA- 5693532). For genes with multiple transcripts, the number of DETs corresponding to each gene is marked by the number brackets. Genes linked to functional terms; Base excision repair (GO:0006284), Base Excision Repair (R-HSA-73884), DNA Double-Strand Break Repair (R-HSA-693532) or Double-strand break repair (GO:0006302), are listed in circles (down-regulated in blue and up-regulated in pink). PBMC; peripheral blood mononuclear cells. Illustration created with BioRender.com. Related to Figure S1.
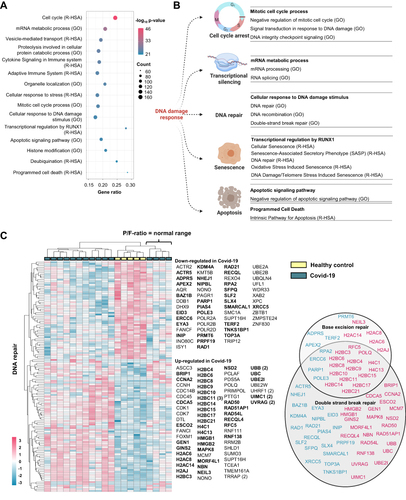
Figure 3 DNA repair gene expression correlates to disease severity. (A) Network plot showing transcripts correlating to P/F-ratio and enriched in DNA repair (GO:0006281). Pearson correlation analysis (p < 0.02, r>±0.6). Genes linked to functional terms; Base excision repair (GO:0006284), Base Excision Repair (R-HSA-73884), DNA Double-Strand Break Repair (R-HSA-693532) or Double-strand break repair (GO:0006302), are listed in circles (down-regulated in blue and up-regulated in pink). (B) Representative correlation plots are shown. MMR; mismatch repair, NER; nucleotide excision repair, BER; base excision repair and DSBR; double strand break repair. Figure is related to Supplemental File 3.
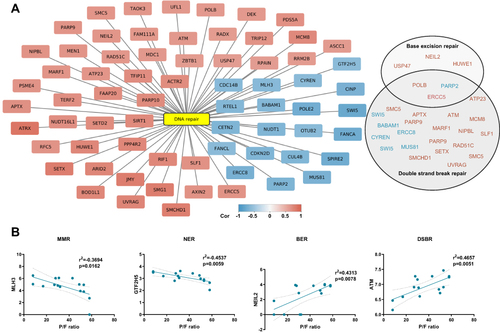
Figure 4 Cellular expression of DNA repair genes in hospitalized Covid-19 patients. Heatmaps generated from publicly available single cell RNA sequencing data (GSE174072) showing genes involved in (A) Base excision repair and (B) Double strand break repair. Hospitalized Covid-19 (n = 7), healthy controls (n = 6).
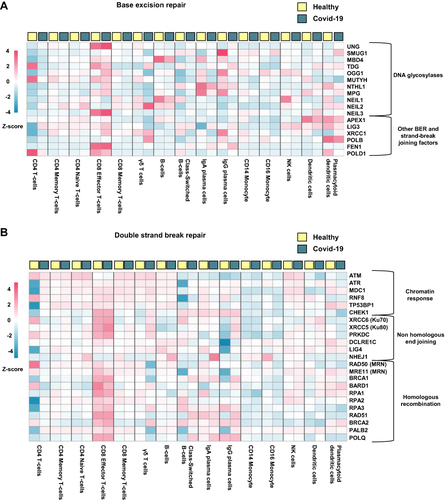
Figure 5 CD4 T cell expression of DNA repair genes in hospitalized Covid-19. Heatmaps generated from publicly available RNA sequencing data (GSE1794478) showing DNA repair genes significantly regulated in CD4 T cells from hospitalized Covid-19 patients compared to control. CD4+CD25− cells were defined as (A) conventional T cells and CD4+CD25highCD127low cells were defined as (B) regulatory T cells.Citation30 DNA repair genes were defined as genes included in gene ontology term DNA repair (GO:0006281) and/or Reactome pathway DNA repair (R-HSA- 5693532). Genes affiliated to functional terms; Base excision repair (GO:0006284), Base Excision Repair (R-HSA-73884), DNA Double-Strand Break Repair (R-HSA-693532) or Double-strand break repair (GO:0006302), are listed in circles (down-regulated in blue and up-regulated in pink). Gene enrichment analysis results are listed in Supplemental File 4).
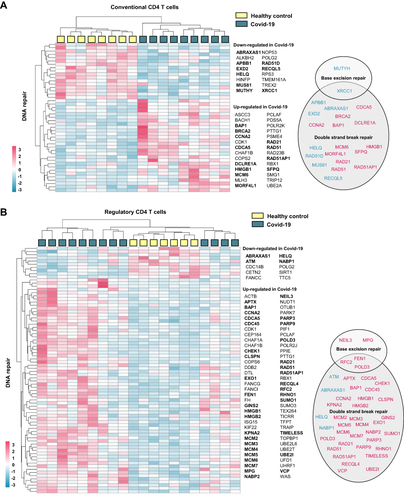
Figure 6 The Hallmark pathway DNA repair is significantly more regulated in immune cells during Covid-19 compared to other lower airway infections. Data are obtained by reanalyzing publicly available data (GSE161731); RNA sequencing of whole blood from patients with Covid-19 (n = 15), lower airway infection due to seasonal coronavirus (n = 51), influenza (n = 17) or bacterial pneumonia (n = 24). (A) Gene Set Enrichment Analysis (GSEA, v.7.4MSigDB) comparing gene expression related to the pathway Hallmark of DNA Repair in; (1) Covid-19 vs bacterial pneumonia, (2) Covid-19 vs acute respiratory infection due to seasonal coronavirus and (3) Covid-19 vs Influenza. (B) Expression levels (log2[FoldChange]) of selected base excision repair (BER) genes and double strand break repair (DSBR) genes in Covid-19 compared to other infections. All selected genes are significantly regulated in Covid-19 vs healthy controls for cohort1 and/or cohort2 and/or CD4 T cells. Student’s t test, *p < 0.05.
![Figure 6 The Hallmark pathway DNA repair is significantly more regulated in immune cells during Covid-19 compared to other lower airway infections. Data are obtained by reanalyzing publicly available data (GSE161731); RNA sequencing of whole blood from patients with Covid-19 (n = 15), lower airway infection due to seasonal coronavirus (n = 51), influenza (n = 17) or bacterial pneumonia (n = 24). (A) Gene Set Enrichment Analysis (GSEA, v.7.4MSigDB) comparing gene expression related to the pathway Hallmark of DNA Repair in; (1) Covid-19 vs bacterial pneumonia, (2) Covid-19 vs acute respiratory infection due to seasonal coronavirus and (3) Covid-19 vs Influenza. (B) Expression levels (log2[FoldChange]) of selected base excision repair (BER) genes and double strand break repair (DSBR) genes in Covid-19 compared to other infections. All selected genes are significantly regulated in Covid-19 vs healthy controls for cohort1 and/or cohort2 and/or CD4 T cells. Student’s t test, *p < 0.05.](/cms/asset/2ae872e2-ae50-431a-95bf-e8ef6827a4f4/djir_a_12152001_f0006_c.jpg)
