Figures & data
Table 1 Primer Sequence for Real-Time PCR
Table 2 Differentially Expressed miRNAs
Figure 1 HK-2 cell cytotoxicity and apoptosis induced by diquat.
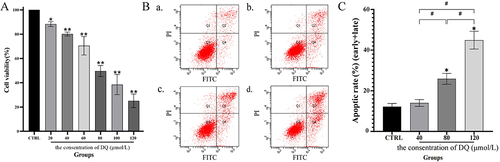
Figure 2 Inter-sample correlation, principal components, and public and unique sequence analysis.
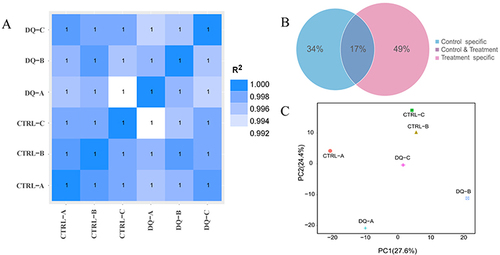
Figure 3 Profiles of the DE miRNAs. HK-2 cells were cultured in a medium containing 80 μmol/L diquat or an equivalent amount of PBS for 24 h.
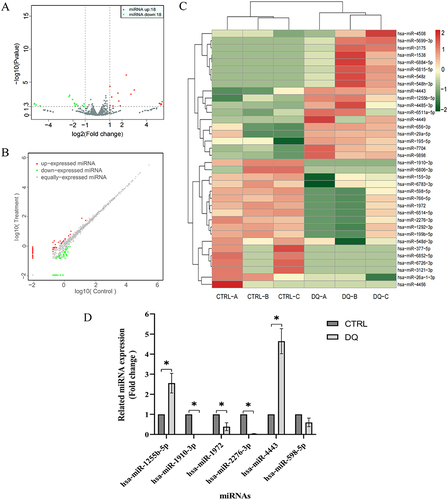
Figure 4 Results of GO and KEGG analyses visualized by imageGP platform.
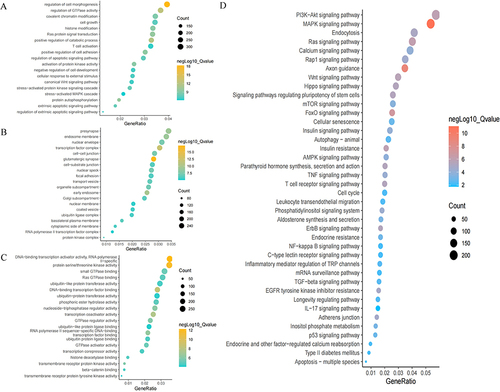
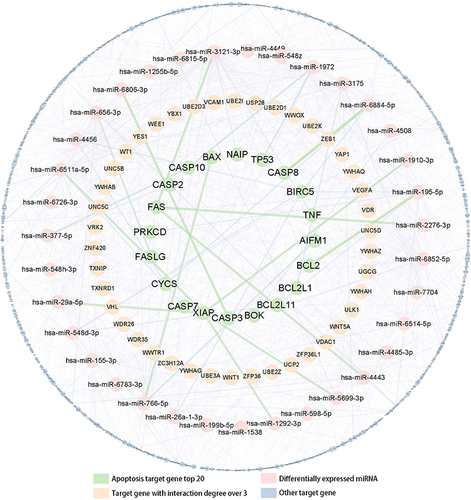
Figure 5 Essential target genes and network regulation map visualized by Cytoscape_v3.6.1 software.
Data Sharing Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
