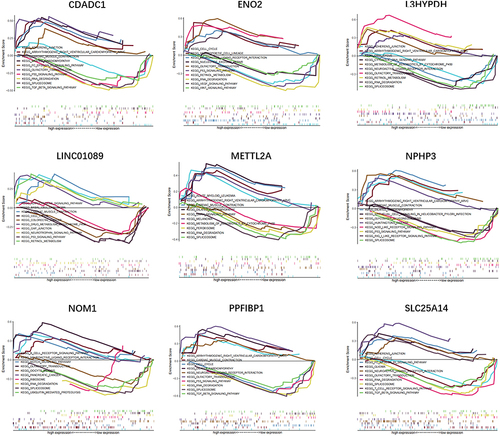
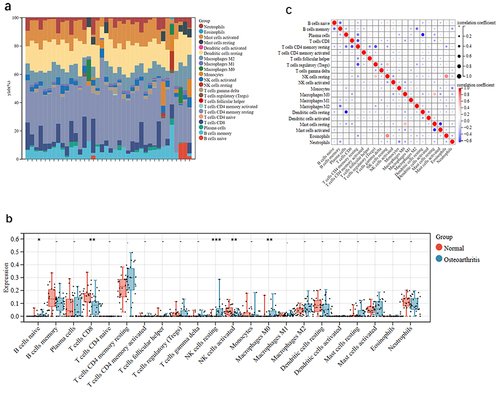
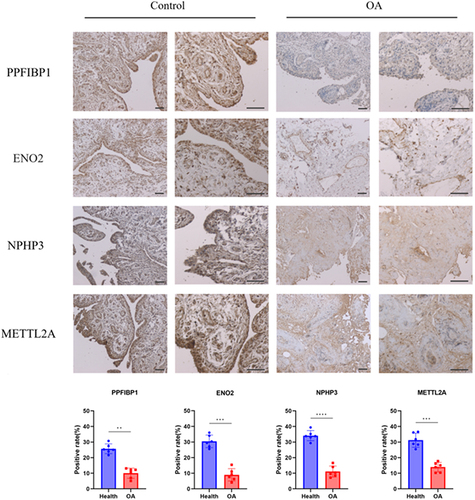
Figures & data
Table 1 Basic Information of Datasets
Figure 2 Differential analysis of genes in OA.
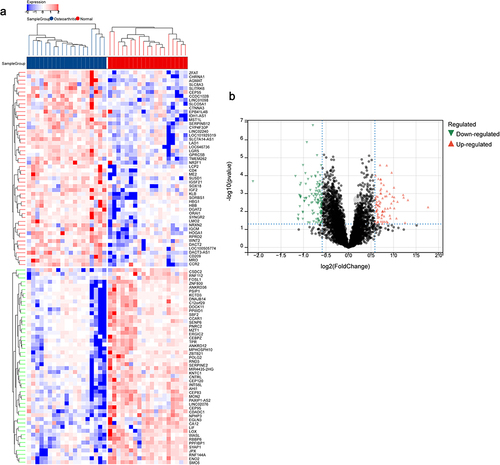
Figure 3 Enrichment analysis of DEGs in OA.
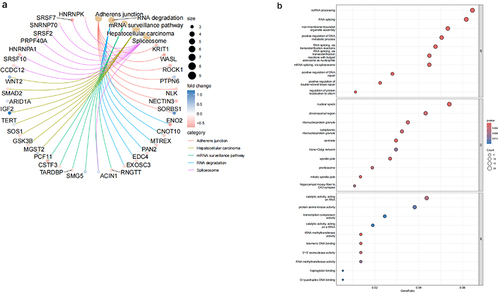
Figure 4 Module genes using WGCNA in OA.
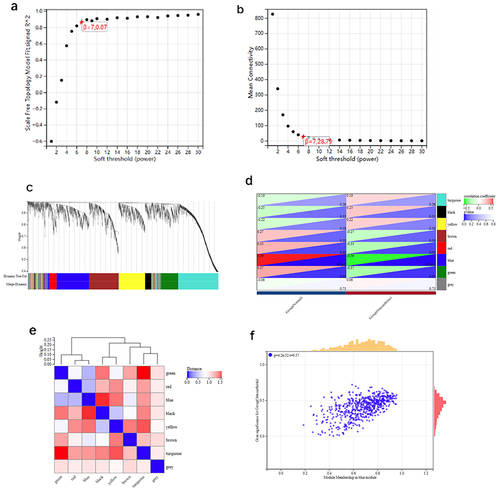
Figure 5 Analysis of the genes of DEGs and key modules.
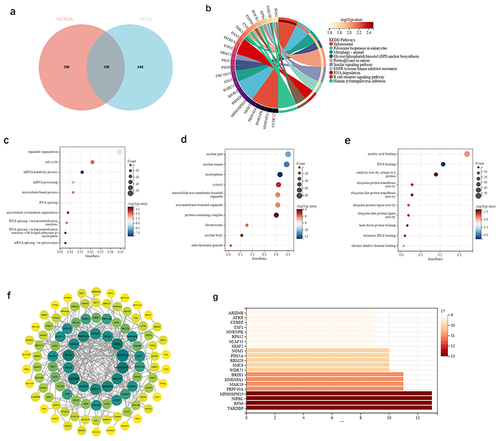
Table 2 Analysis Results of Machine Learning
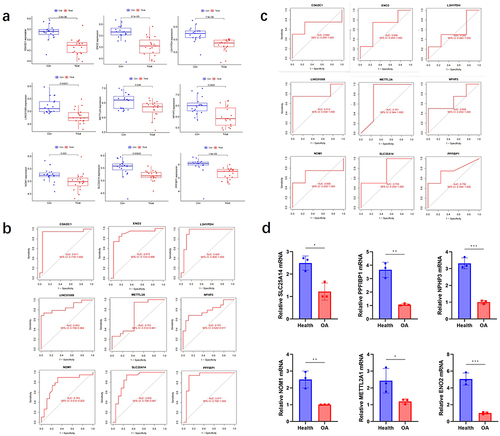
Figure 6 Machine learning was used to screen the diagnostic genes of OA.
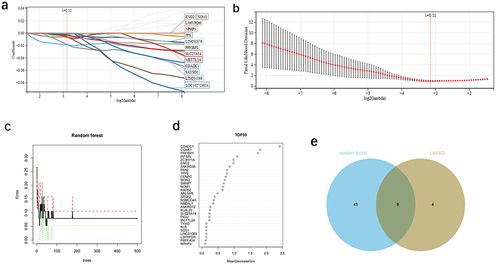
Figure 7 The model genes expression.