Figures & data
Table 1 Abbreviation, botanical name, used part, and yield of extract of each screened plant
Table 2 Primer pair sets and parameters used in the real time polymerase chain reaction analysis
Table 3 messengerRNA expression of TNFα, IL1β, NFκB1, NFκB2, NOS2, COX2, NFE2L2, and PPARγ in RAW264.7 cells treated with 50, 100, and 200 μM of H2O2 for 30 and 60 minutes in comparison to control cells
Figure 1 messengerRNA expression of TNFα, IL1β, COX2, NOS2, NFκB1, NFκB2, NFE2L2, and PPARγ in RAW264.7 cells treated with 10 μg/ml of each extract for 24 hours after a pre-stimulation using 200 μM H2O2 for 1 hour relative to H2O2-treated cells.
Notes: (A) TNFα; (B) IL1β; (C) COX2; (D) NOS2; (E) NFκB1; (F) NFκB2; (G) NFE2L2; (H) PPARγ. To compare the bioactivities between six plant extracts for each gene, the general linear model was used with fixed effect for extracts. Bars represent the log2(n-fold) mean ± standard error (n=4). The effect of plant extracts for each gene are indicated with different capital letters denoting significant differences at P<0.001.
Abbreviations: AL, Arctium lappa; CS, Camellia sinensis; EA, Echinacea angustifolia; ES, Eleutherococcus senticosus; PG, Panax ginseng; VM, Vaccinium myrtillus; COX2, cyclooxygenase 2; IL1β, interleukin 1β ; NFE2L2, nuclear factor E2-related factor 2; NFκB, nuclear factor κB; NOS2, nitric oxide synthase 2; PPARγ, proliferator-activated receptor γ; TNFα, tumor necrosis factor α.
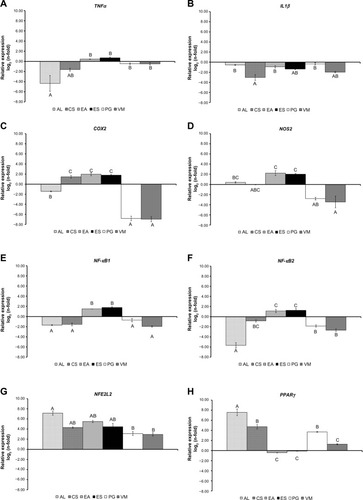
Table 4 Cell viability of RAW264.7 cells treated with 50, 100, and 200 μg/ml of H2O2 for 30 and 60 minutes
Table 5 Cell viability of RAW264.7 cells treated with 1, 10, 100, and 200 μg/ml plant extracts for 24 hours
