Figures & data
Table 1 Top ten up- and downregulated DElncRNAs between PD and normal controls
Table 2 Top ten up- and downregulated DEmRNAs between PD and normal controls
Figure 1 DElncRNAs and DEmRNAs between PD and normal controls.
Abbreviations: DElncRNAs, differentially expressed long noncoding RNAs; DEmRNAs, differentially expressed micro RNAs; PD, Parkinson’s disease.
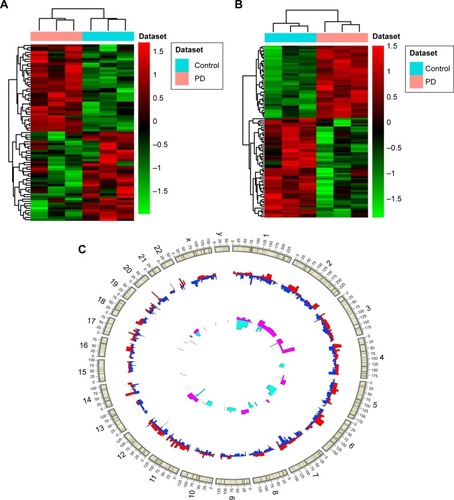
Figure 2 Significantly enriched GO terms and KEGG pathways of DEmRNAs between PD and normal controls.
Abbreviations: BP, biological process; CC, cellular component; DEmRNAs, differentially expressed mRNAs; FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function.
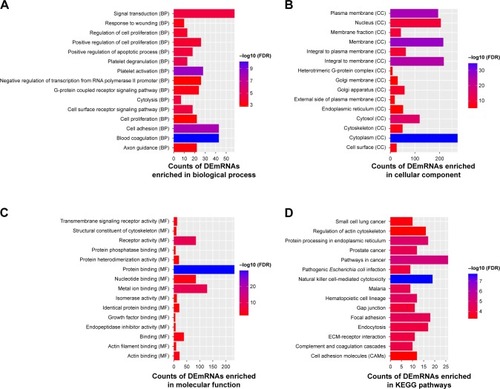
Figure 3 PD-specific PPI network.
Abbreviations: DEmRNAs, differentially expressed mRNAs; PD, Parkinson’s disease; PPI, protein–protein interaction.
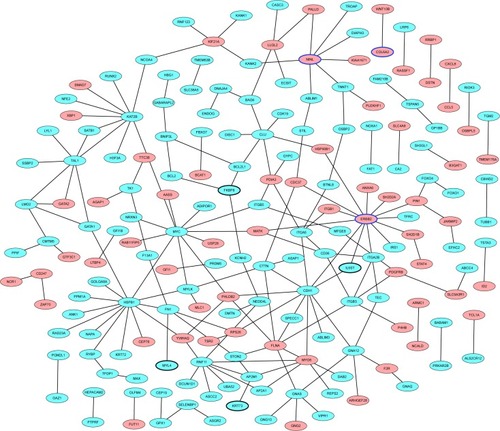
Table 3 Nearby targeted DEmRNAs of DElncRNAs between PD and normal controls
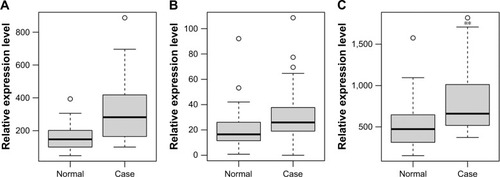
Figure 4 Validation of selected DEmRNAs in GSE57475.
Abbreviations: DEmRNAs, differentially expressed micro RNAs; PD, Parkinson’s disease.
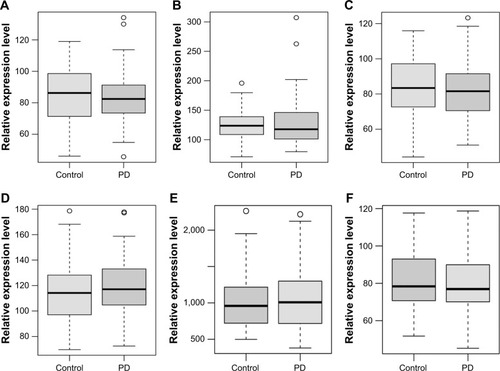
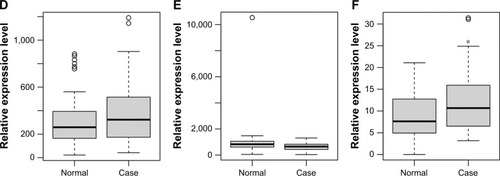
Figure 5 Validation of selected DEmRNAs in GSE68719.
Abbreviations: DEmRNAs, differentially expressed micro RNAs; PD, Parkinson’s disease.