Figures & data
Figure 1 Illustration of the methodology followed in the discovery phase of the experiments. Florescent labeling of proteins isolated from clinical phenotypes of schizophrenia and Parkinson’s disease was done with Cyanine 3 (green) and Cyanine 5 (red). Internal standard was labeled with Cyanine 2 (blue). Combined image after DIGE is shown under gels. Relative quantitation and consistency of expression were analyzed by DeCyder software. Differentially expressed spots were picked from preparative gels. Spots were digested with trypsin. Mass spectrometry analysis was done for protein identification.
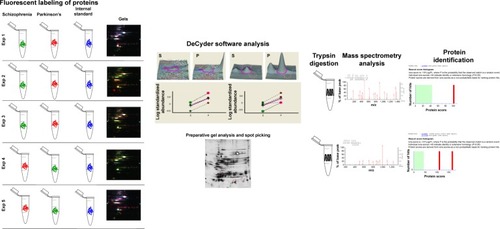
Table 1 Clinical profile of patients used in DIGE experiment
Table 2 Demographic profile of patients recruited for ELISA
Table 3 Clinical profile of patients receiving pharmacological therapy and showing side effects
Figure 2 Differential gel electrophoretic gel images show differential protein expression between the serum of patients with Parkinson’s disease and schizophrenia. A total of five biological replicate gels were run using five sets of serum samples from patients with Parkinson’s disease and schizophrenia. Spots that equally expressed in both clinical phenotypes are indicated by white arrows. Spots that are differentially expressed are indicated by red or green arrows. Spots that are differentially expressed, with similar relative ratios and consistently present across all the five experiments, are indicated by a blue box.
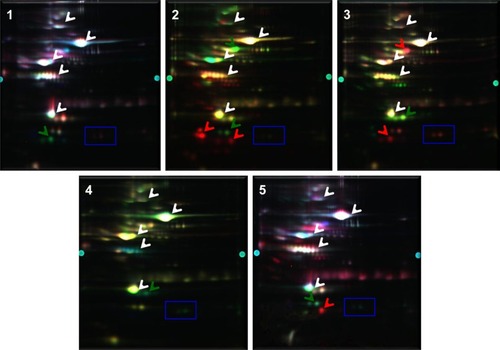
Figure 3 Magnified view of the gels showing the varying expression of proteins in patients with Parkinson’s disease and schizophrenia. Images correspond to the two phenotypes from five biological replicate gels. The protein spots that were picked for identification are indicated by black arrows.
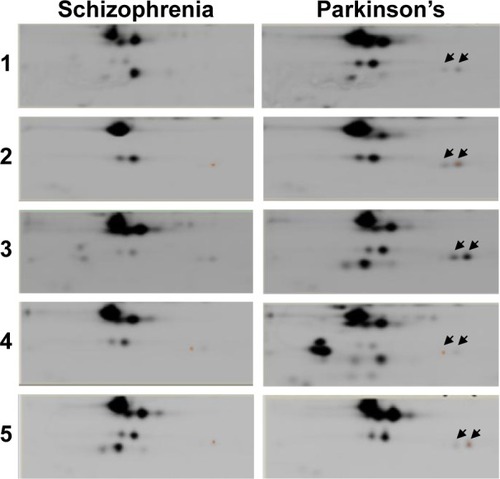
Figure 4 Relative abundance of the differentially expressed proteins that are marked. Top panel shows spot intensities in terms of peak volume ratios (±3.0) based on DeCyder software analysis. Graphical representation of protein spots differentially expressed in serum of patients with Parkinson’s disease (P) as compared to schizophrenia (S) (P<0.05). Spot data from same gels are connected by dotted lines.
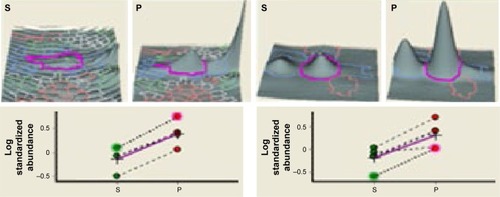
Table 4 Profile of identified protein spots
Table 5 Pearson correlation between α-globin concentrations and disease scoring, gender, and age
Table 6 Pearson correlation between β-globin concentrations and disease scoring, gender, and age
Figure 5 Expression of α- and β-globins in the serum of patients with Parkinson’s disease and schizophrenia analyzed by ELISA. Clinical phenotypes comprise of Parkinson’s disease naïve, Parkinson’s treated, Parkinson’s treated with schizophrenic side effects, schizophrenia treated with Parkinson’s disease-like side effects, schizophrenia treated and schizophrenia naïve patients. Mean ± standard error of mean of the values is shown by horizontal lines. The bars represent the concentrations as the average of duplicate readings of each patient sample. Trend lines of α-globin (y =-1.12x+16.4; R2 =0.86) and β-globins (y =-0.85x+14.2; R2 =0.81) across the six clinical phenotypes are shown in blue dotted lines in (A and B), respectively. Diagrammatic representation of the dopamine concentration in cerebrospinal fluid (CSF) is shown along the x-axis.Citation47,Citation48 *Statistical significance with P<0.05.
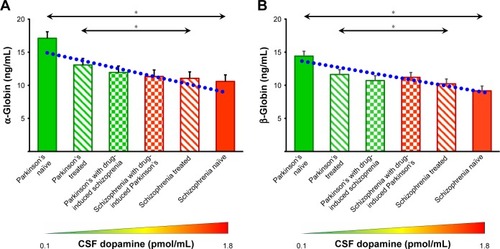
Figure 6 Correlation analysis of α- and β-globins expression with duration of pharmacotherapy. “r” represents the correlation coefficient.
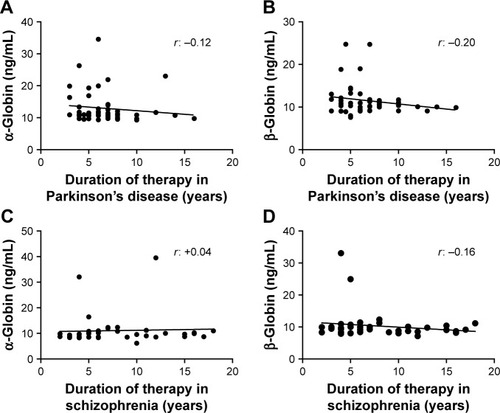
Figure 7 Western blot analysis of bisphosphoglycerate mutase. P1–P4 show four representative samples from Parkinson’s disease group; S1–S4 show four representative samples from schizophrenia group. +, Positive control; M, marker.

Table 7 Pharmacotherapeutic monitoring value of α-globin and β-globin to differentiate Parkinson’s disease and schizophrenia
Figure S1 SDS-PAGE showing removal of the high abundant proteins, albumin and immunoglobulin heavy chain, from serum samples of patients with Parkinson’s disease (P) and schizophrenia (S).
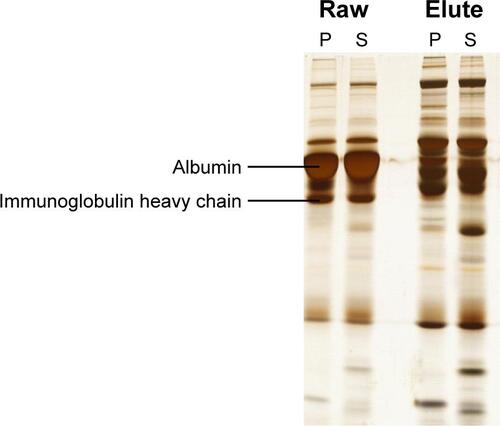
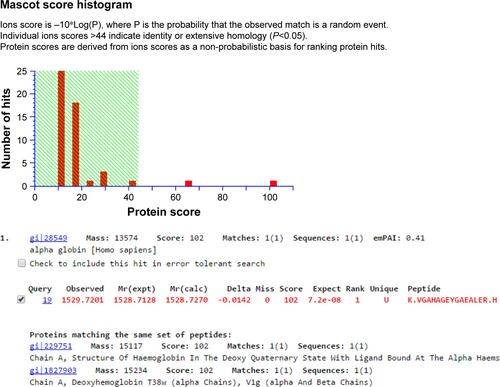
Figure S2 Snapshot of mass spectrometry data analysis of spot number 1,570 for protein identification using MASCOT software.
Figure S3 Snapshot of mass spectrometry data analysis of spot number 1,527 for protein identification using MASCOT software.