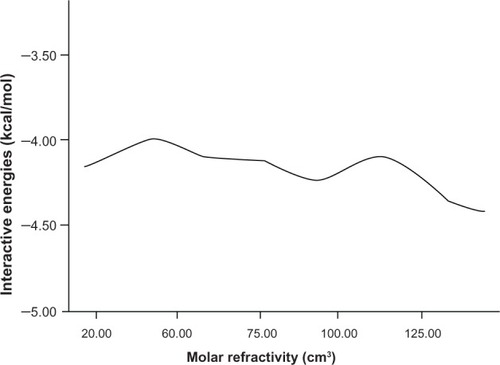
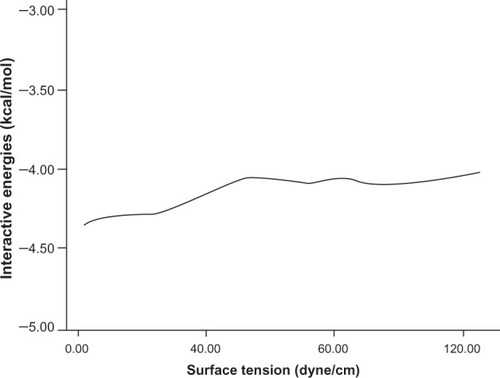
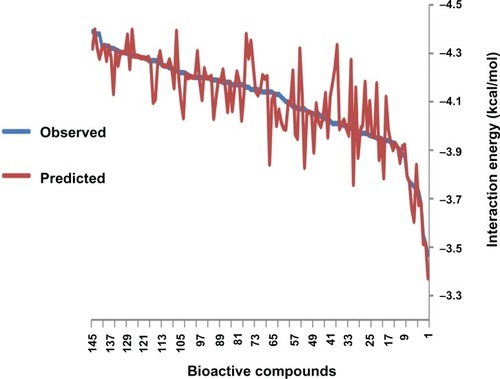
Figures & data
Figure 1 One molecule loaded in silicon-nanostructure (before minimization). The box inside silicon nanochambers shows the typical docking space.
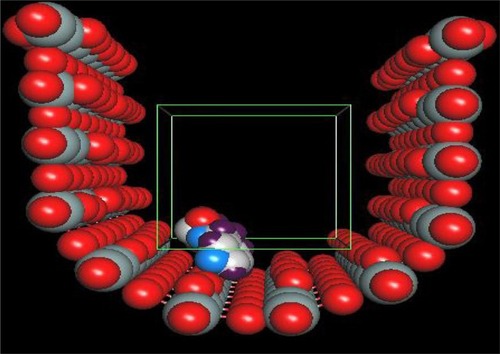
Table 2 The neural network architectures produced with the descriptors for bio-active compound with the error limit of <0.05
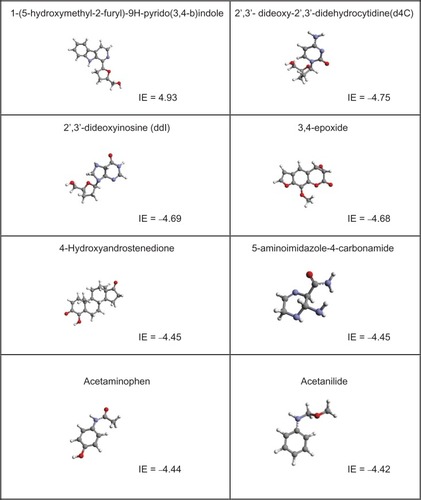
Table 1 Names of drugs and bioactive molecules used in this study