Figures & data
Figure 1 The expression of RANKL in GC tissues.
Abbreviation: GC, gastric cancer.
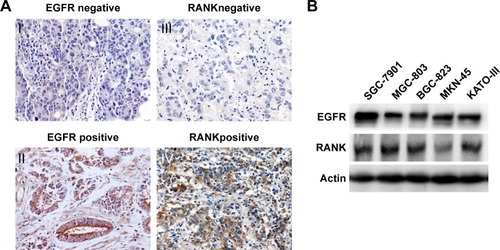
Table 1 Correlation between the expression of EGFR or RANK and the clinicopathological factors in primary GC patients
Table 2 Spearman’s correlations between EGFR and RANK expression in primary GC patients
Figure 2 RANKL stimulated EGFR phosphorylation and downstream signaling in GC cells.
Abbreviation: GC, gastric cancer.
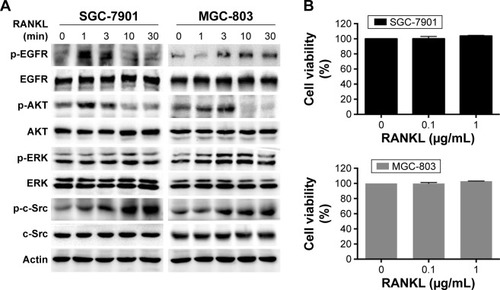
Figure 3 The effect of RANKL on sensitivity to cetuximab in GC cells.
Abbreviation: GC, gastric cancer.
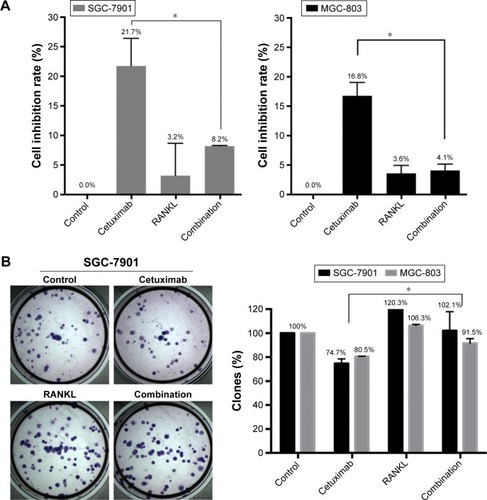
Figure 4 The effect of RANKL/RANK pathway on cetuximab sensitivity in SGC-7901 cells.
Abbreviations: NS, non-silencing; siRNA, small interfering RNA.
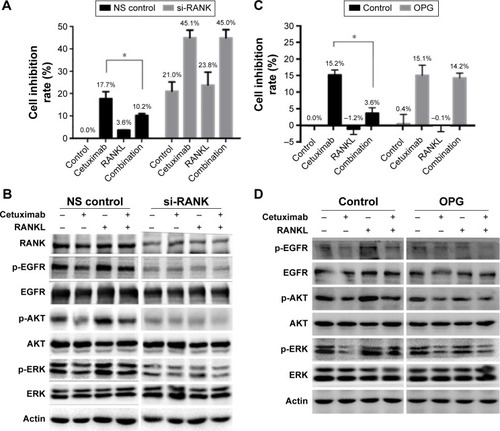
Figure 5 The effect of dasatinib on RANKL-induced cetuximab resistance in SGC-7901 cells.
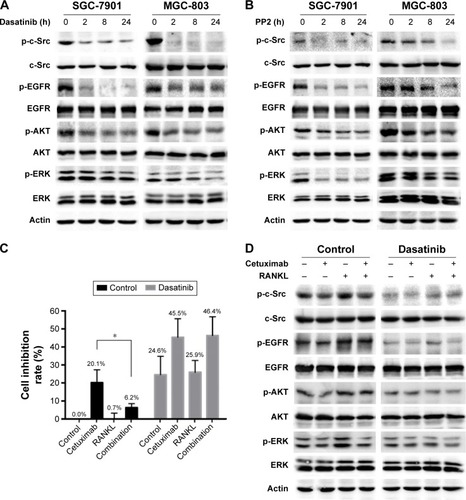
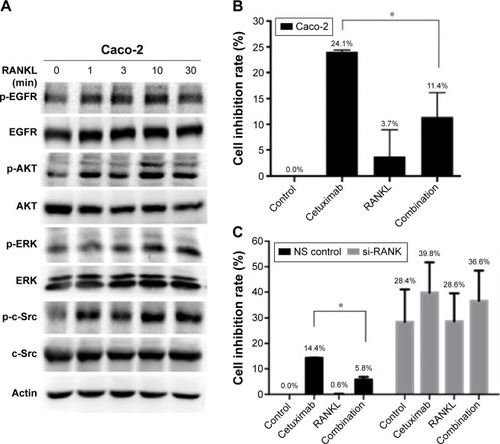
Figure 6 The effect of RANKL/RANK pathway on sensitivity to cetuximab in Caco-2 cells.
Abbreviations: NS, non-silencing; siRNA, small interfering RNA.