Figures & data
Table 1 Primers used in qRT-PCR analysis
Figure 1 ARHGAP1 expression in cervical cancer tissues and their adjacent normal tissues.
Abbreviation: TCGA, The Cancer Genome Atlas.
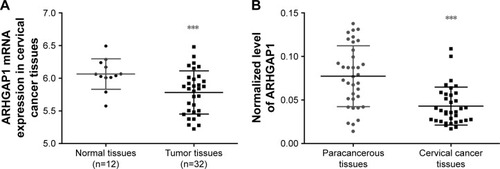
Table 2 Correlation between the expression of ARHGAP1 and clinicopathologic features
Figure 2 ARHGAP1 expression in cervical carcinoma cell lines.
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; lenti-ARHGAP1, lentivirus targeting ARHGAP1; NC, negative control; qRT-PCR, quantitative real-time polymerase chain reaction; shNC, shRNA-NC.
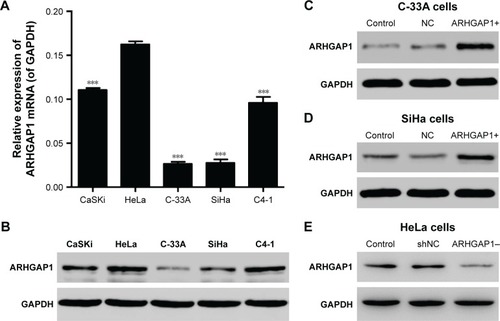
Figure 3 Cell survival was assessed by CCK-8 assay.
Abbreviations: CCK-8, Cell Counting Kit-8; OD, optical density; lenti-ARHGAP1, lentivirus targeting ARHGAP1; NC, negative control; shNC, shRNA-NC.
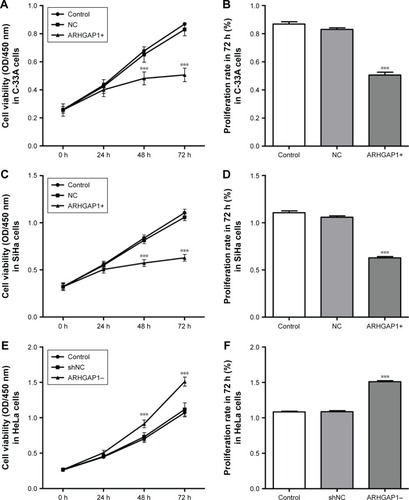
Figure 4 Effects of ARHGAP1 on migration of both C-33A and SiHa cells.
Abbreviations: NC, negative control; shNC, shRNA-NC.
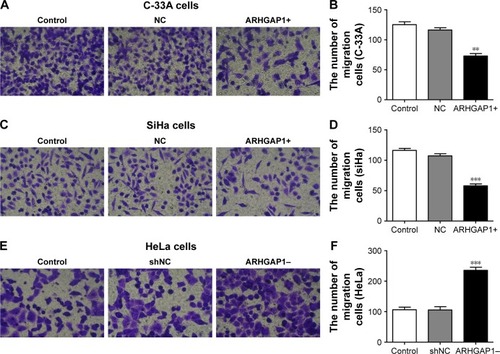
Figure 5 Effects of ARHGAP1 on invasion of C-33A and SiHa cells.
Abbreviations: lenti-ARHGAP1, lentivirus targeting ARHGAP1; NC, negative control; shNC, shRNA-NC.
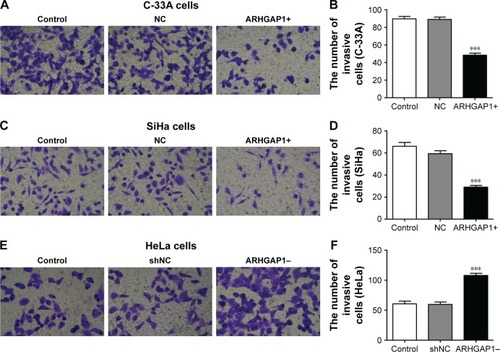
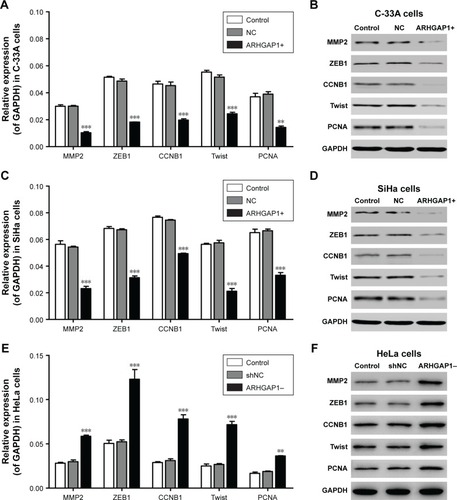
Figure 6 Effects of ARHGAP1 on the protein expressions of MMP2, ZEB1, CCNB1, Twist and PCNA in C-33A and SiHa cells.
Abbreviations: CCNB1, Cyclin B1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MMP2, matrix metallopeptidase 2; PCNA, proliferating cell nuclear antigen; NC, negative control; shNC, shRNA-NC; Twist, twist family bHLH transcription factor 1; ZEB1, zinc finger E-box binding homeobox 1.