Figures & data
Figure 1 Upregulation of CRNDE was observed in both colorectal cancer tissues and cells lines. (A) CRNDE was significantly upregulated in colorectal cancer tissues in tumor tissues relative to adjacent normal tissues (n=10). (B) Expression of CRNDE in colorectal cancer cell lines (SW480, HCT116 and HT-29) and normal human colon cell lines (HcoEpic and NCM460). Data represent three independent experiments (average and sem of triplicate samples). **P<0.01.
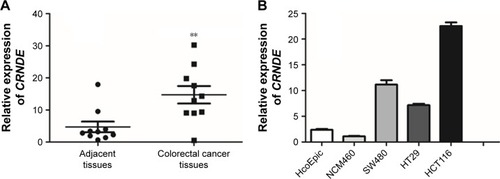
Figure 2 Knockdown of CRNDE inhibited the proliferation, migration and invasion of colorectal cancer cells. (A) The CRNDE level of SW480 and HCT116 cells was decreased after being transfected with CRNDE siRNAs or si-control control. (B) Cell proliferation analysis using CCK-8 assay after being transfected with CRNDE siRNAs or si-control. (C) Scratch-wound healing assay was used to assess the migration potency of HCT116 cells after being transfected with CRNDE siRNAs or si-control control. The wound closure was calculated at 48 hours. (D, E) Matrigel invasion assay was performed to test the change of invasion potency after being transfected with CRNDE siRNAs or si-control control. Representative images of the invaded cells are shown. Six fields for each chamber were photographed using an inverted microscope and camera, and invading cells counted in each field. The average number was shown. (F, G) Flow cytometry was used to evaluate the cell apoptosis of HCT116 cells with knockdown of CRNDE. Data represent three independent experiments (average and sem of triplicate samples). *P<0.05, **P<0.01, ***P<0.001.
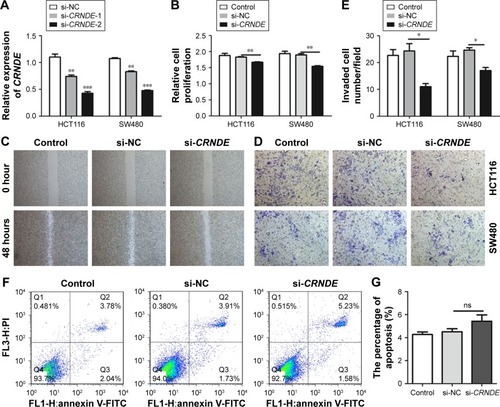
Figure 3 Overexpression of CRNDE enhances the proliferation, migration and invasion of colorectal cancer cells. HCT116 cells were transfected with pcDNA-CRNDE or pcDNA-Vector. (A) CCK-8 assay was performed to assess cell viability. (B) Scratch-wound healing assay was used to assess the migration potency of HCT116 cells after being transfected. (C, D) Matrigel invasion assay was performed to test the change of invasion potency after being transfected. Representative images of the invaded cells are shown. Six fields for each chamber were photographed using an inverted microscope and camera, and invading cells counted in each field. The average number is shown. (E) Flow cytometry was used to evaluate the cell apoptosis. Data represent three independent experiments (average and sem of triplicate samples). *P<0.05, **P<0.01.
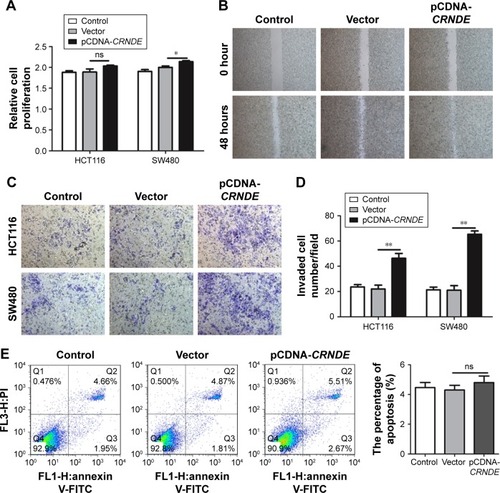
Figure 4 Knockdown of CRNDE enhances colorectal cancer cell sensitivity to OXA. HCT116 cells were transfected with CRNDE siRNAs or si-control control. After 48 hours, cells were treated with OXA. (A) The transfected HCT116 cells were treated with 15 μM (50% inhibitory concentration values) OXA, and CCK-8 assay was performed to assess cell viability after 0, 12, 24, and 48 hours. (B) The transfected HCT116 cells were treated with different doses of OXA, and CCK-8 assay was performed to assess cell viability after 48 hours. (C) Western blot was used to analyze the protein expression of γH2AX in HCT116 cells after 48 hours of OXA treatment. (D) Densitometry plot of results from . The relative expression levels were normalized to GAPDH. (E, F) Flow cytometric analysis was used to evaluate the apoptosis of HCT116 cells after 48 hours of OXA treatment. Data represent three independent experiments (average and sem of triplicate samples). *P<0.05, **P<0.01.
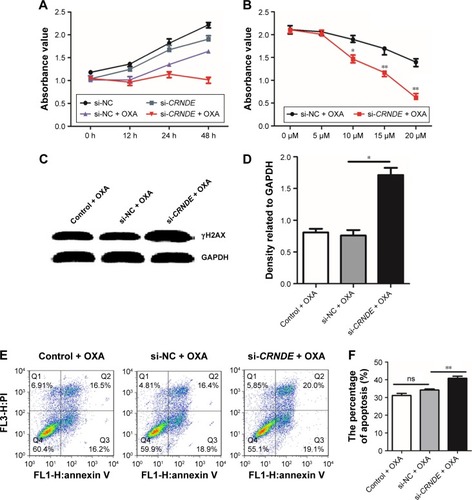
Figure 5 Overexpression of CRNDE enhances the OXA chemoresistance of colorectal cancer cells. HCT116 cells were transfected with pcDNA-CRNDE or pcDNA-Vector. After 48 hours, cells were treated with 15 μM (50% inhibitory concentration values) OXA for 24 hours or 48 hours. (A) CCK-8 assay was performed after 24 hours of OXA treatment. (B) Western blot was used to analyze the protein expression of γH2AX in HCT116 cells after 48 hours of OXA treatment. (C, D) Flow cytometric analysis was used to evaluate the apoptosis of HCT116 cells after 48 hours of OXA treatment. Data represent three independent experiments (average and sem of triplicate samples). *P<0.05, **P<0.01, ***P<0.001.
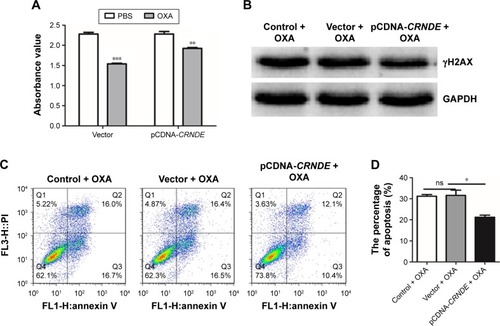
Figure 6 CRNDE is a target of miR-136 and modulates its target E2F1. (A) Schematic diagrams of the mutual interactions between miR-136 and CRNDE. (B) The inverse correlation of the expression of CRNDE and miR-136 among the tissue samples was measured by linear regression analysis. (C) CRNDE levels in the immunoprecipitates were measured by Q-PCR and presented as fold enrichment in Ago2 relative to IgG immunoprecipitates. (D) The luciferase reporter plasmid containing wild-type or mutant CRNDE was co-transfected into HEK293T cells with pre-miR-136 mimics or pre-NC. Luciferase reporter genes assay was performed to determine if the luciferase activity was normalized to Renilla activity. Three independent experiments were performed using one-way ANOVA analysis. (E) The predicted miR-136 binding sites in the 3-UTR of E2F1 are showed in schematic diagrams. (F) The gene level of E2F1 in HCT116 cells transfected with pre-miR-136 and pre-control. (G) The protein level of E2F1 in HCT116 cells transfected with pre-miR-136 and pre-control. (H) The HCT116 cells were transfected with pcDNA-Vector, pCDNA-CRNDE or pCDNA-CRNDE-mut (miR-136 and CRNDE binding sites were mutated) plasmid and the gene of E2F1 was tested. (I) The HCT116 cells were transfected with pcDNA-Vector, pCDNA-CRNDE or pCDNA-CRNDE-mut (miR-136 and CRNDE binding sites were mutated) plasmid and protein levels of E2F1 were tested. Data represent three independent experiments (average and sem of triplicate samples). **P<0.01, ***P<0.001.
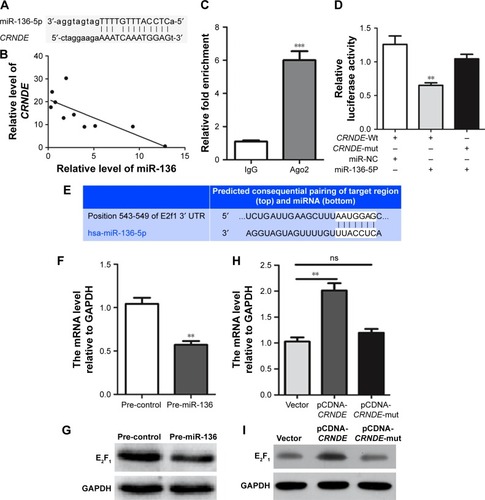
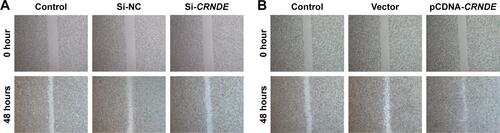
Figure S1 CRNDE was involved in cell migration of colorectal cancer cells. (A) Scratch-wound healing assay was used to assess the migration potency of SW480 cells after being transfected with CRNDE siRNAs or si-control control. The wound closure was calculated at 48 hours. (B) Scratch-wound healing assay was used to assess the migration potency of SW480 cells after being transfected with pcDNA-CRNDE or pcDNA-Vector. The wound closure was calculated at 48 hours.