Figures & data
Table 1 Association between CUL4A expression and clinicopathologic features of colorectal cancer patients
Figure 1 CUL4A is overexpressed and associated with prognosis in colorectal cancer.
Abbreviation: qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
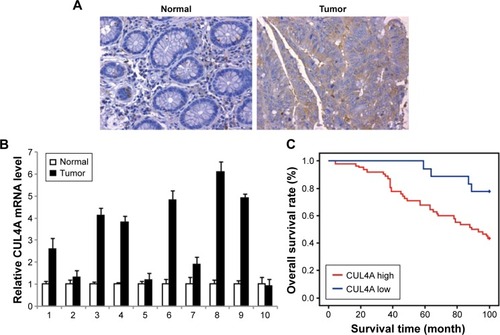
Figure 2 CUL4A promotes colorectal cancer cell line HCT-116 growth and migration.
Abbreviations: MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
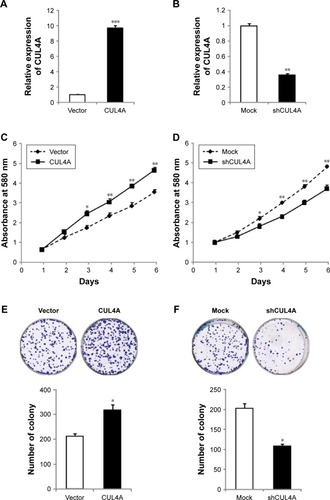
Figure 3 CUL4A promotes migration of colorectal cancer cell line HCT-116.
Abbreviations: SD, standard deviation; h, hours.
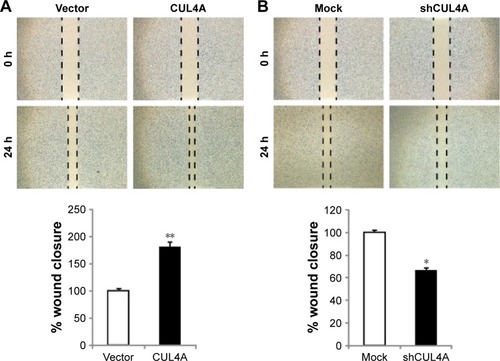
Figure 4 CUL4A promotes epithelial–mesenchymal transition by changing the H3K4me3 level in promoter regions of E-cadherin, N-cadherin and vimentin.
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IgG, immunoglobulin G; H3K4me3, H3K4 trimethylation; PCR, polymerase chain reaction; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
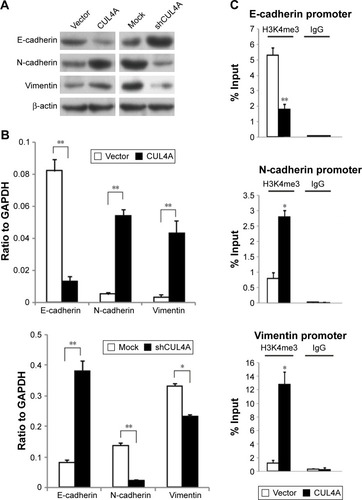
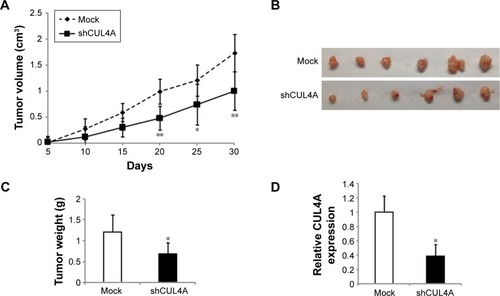
Figure 5 Knockdown of CUL4A suppresses tumor growth in vivo.
Abbreviations: qPCR, quantitative polymerase chain reaction; SD, standard deviation.