Figures & data
Table 1 Primer sequences used for qRT-PCR
Figure 1 miR-320a relative expression in HCC samples and cell lines.
Abbreviations: HCC, hepatocellular carcinoma; miR-320a, microRNA-320a; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; ns, nonsignificant.
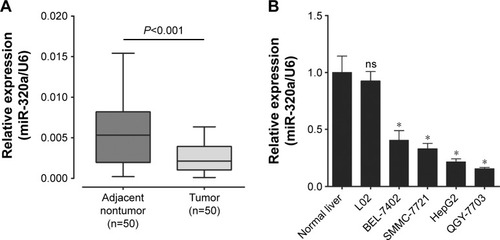
Figure 2 miR-320a directly binds to the coding DNA sequence of c-Myc.
Abbreviations: CDS, coding DNA sequence; DNA, deoxyribonucleic acid; NC, negative control; miR-320a, microRNA-320a; UTR, untranslated regions.
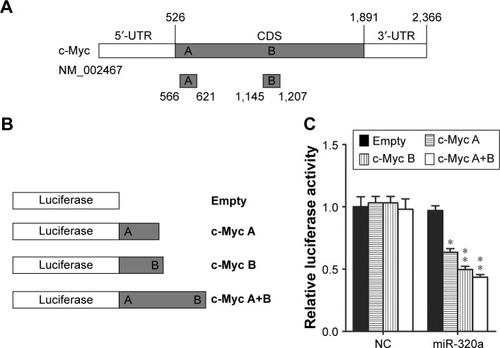
Figure 3 miR-320a downregulates endogenous c-Myc expression in HCC cells.
Abbreviations: HCC, hepatocellular carcinoma; IHC, immunohistochemistry; miR-320a, microRNA-320a; NC, negative control; qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
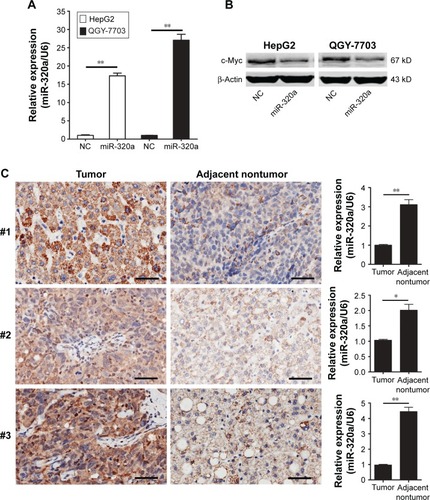
Figure 4 Upregulation of miR-320a inhibited c-Myc expression and hepatocellular carcinoma cells proliferation by targeting c-Myc in vitro.
Abbreviations: miR-320a, microRNA-320a; NC, negative control; OD, optical density; si or siRNA, small interfering RNA.
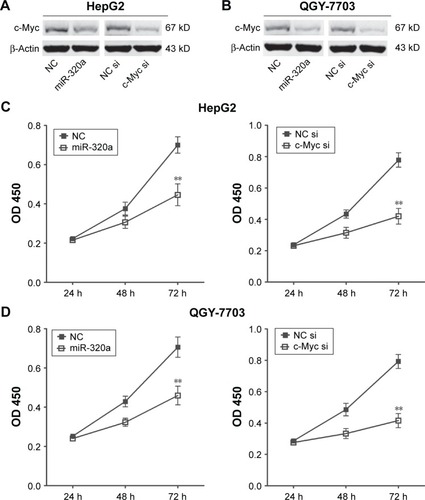
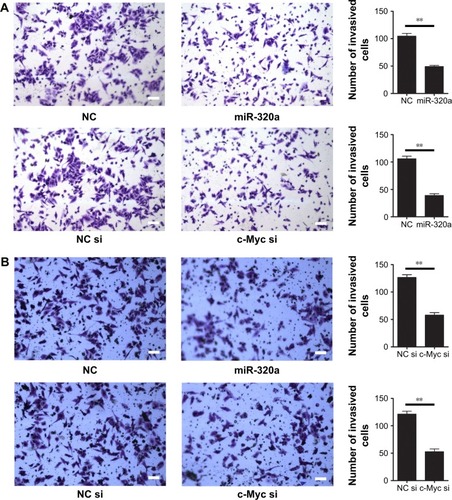
Figure 5 Upregulation of miR-320a inhibited hepatocellular carcinoma cells invasion by targeting c-Myc in vitro.
Abbreviations: miR-320a, microRNA-320a; NC, negative control; si or siRNA, small interfering RNA.