Figures & data
Figure 1 SCC9 and SCC25 after exposure to nimesulide and cisplatin: induction of apoptosis (SCC9: A, SCC25: B) and necrosis (SCC9: C, SCC25: D) is detected by Annexin-V and 7-AAD using flow cytometry, respectively. Adenine nucleotide contents of SCC9 and SCC25 cells were measured after 48-h drug exposure. ATP and energy charge of SCC9 (E, F) and SCC25 (G, H) cells are illustrated. *P<0.05 versus control group indicates statistical significance. Data are mean ± SD of values from at least three experiments for each treatment. Line 1: control; line 2: cisplatin; lane 3: nimesulide, and line 4: nimesulide–cisplatin.
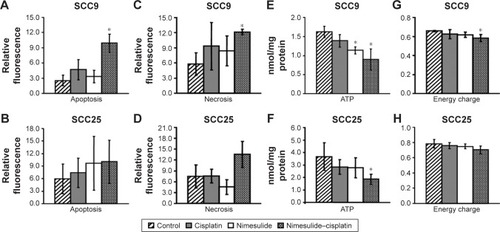
Figure 2 Immunohistochemical detection of apoptotic cells using M30 CytoDeath antibody in SCC9 and SCC25 cells. After 48 h, control SCC25 cells show only slight staining (A), whereas an enrichment of cytoskeletal staining is observed, together with a slight decrease in cell number, after treatment with 0.5 µmol/L cisplatin (B). Treatment with 100 µmol/L nimesulide alone (C) or in combination with 0.5 µmol/L cisplatin (D) reduced the SCC25 cell number and increased the number of apoptotic cells (clear enrichment of cytoskeletal staining). D contains a collage of apoptotic cells after incubation with nimesulide/cisplatin.
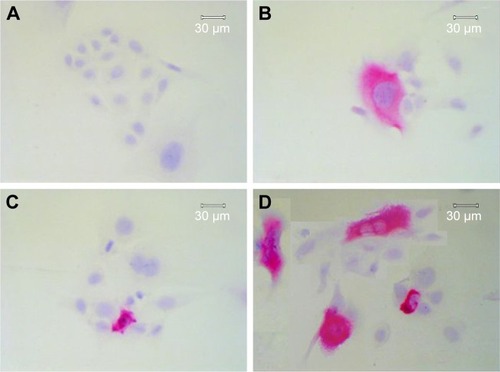
Figure 3 Proteomics data supports histone regulation by combination therapy. MS/MS spectra are shown for two iTRAQ-labeled peptides each from histones H2A and H2B, respectively. All peptides were identified with high (99%) confidence with ProteinPilot software. Tables illustrate b- and y-ions matched with experimental data. iTRAQ reporter ions are highlighted in the raw spectra panes. Reporter peaks at m/z =114.1, reflecting combination therapy are considerably more prominent than reporter peaks reflecting single-drug therapy (m/z =115.1 nimesulide, m/z =116.1 cisplatin) or control samples (m/z =117.1). Regulatory ratios were calculated with ProteinPilot software from raw reporter areas, properly accounting for isotope correction and normalization. As nuclei were removed by a centrifugation step during the experimental workflow, ratios observed in the shotgun proteomics experiment should reflect the relative abundance of proteins in the cytosol. We interpret the increased relative ratio of H2A and H2B in the combination therapy sample as further support for increased apoptosis in combination-treated cells, possibly leading to increased release of these histones from nuclei into the cytosolic environment.
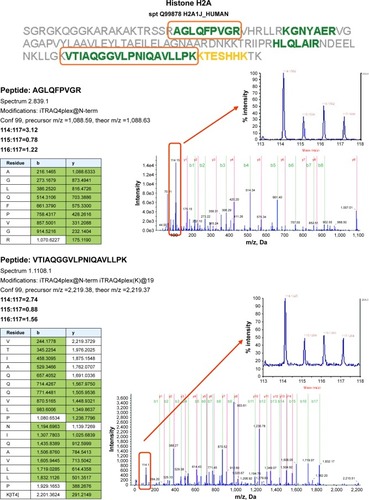
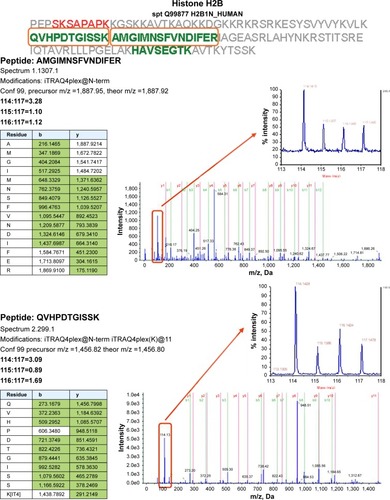
Table 1 List of regulated proteins after exposure to nimesulide and cisplatin
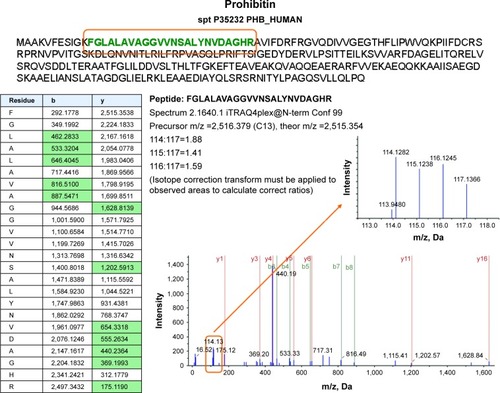
Figure 4 Proteome analysis shows a slight regulation of prohibitin, particularly after combined treatment with nimesulide and cisplatin. MS/MS spectra are shown for two iTRAQ-labeled peptides from prohibitin with high (99%) confidence, using ProteinPilot software. Reporter peaks at m/z =114.1 reflecting combination therapy are considerably more prominent than reporter peaks reflecting single-drug therapy (m/z =115.1 nimesulide, m/z =116.1 cisplatin) or control samples (m/z =117.1).