Figures & data
Figure 1 The IHC of normal liver tissues.
Abbreviation: IHC, immunohistochemistry.
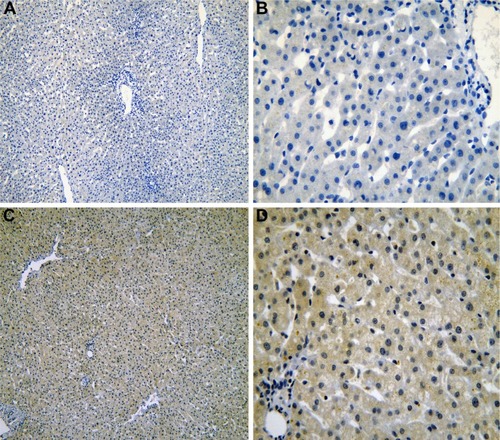
Figure 2 The IHC of liver tissues with cirrhosis.
Abbreviation: IHC, immunohistochemistry.
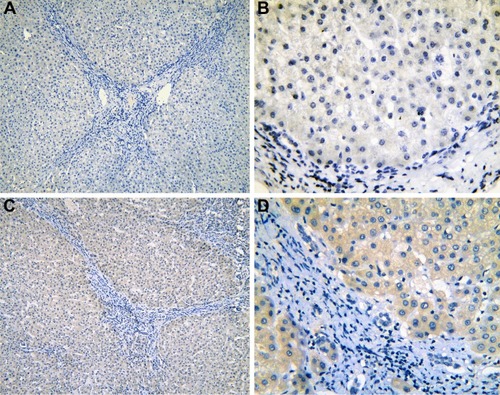
Figure 3 The IHC of para-HCC tissues with cirrhosis.
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
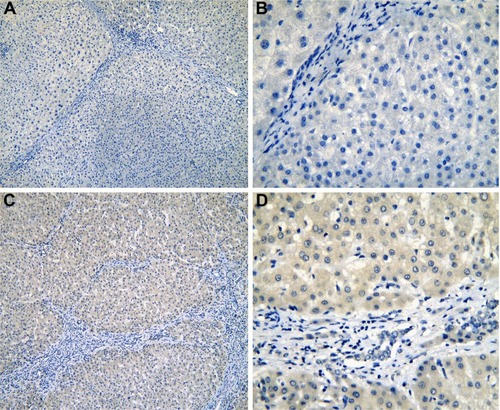
Figure 4 The IHC of normal liver tissues (para-HCC).
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
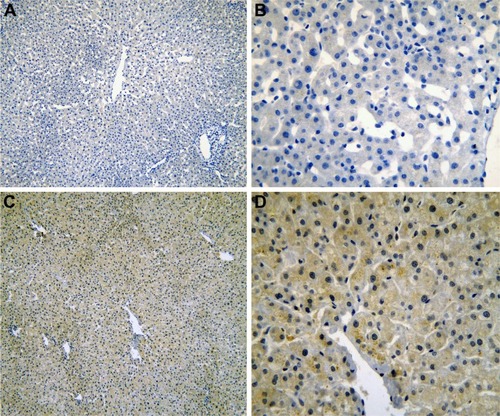
Figure 5 The IHC of HCC tissues.
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
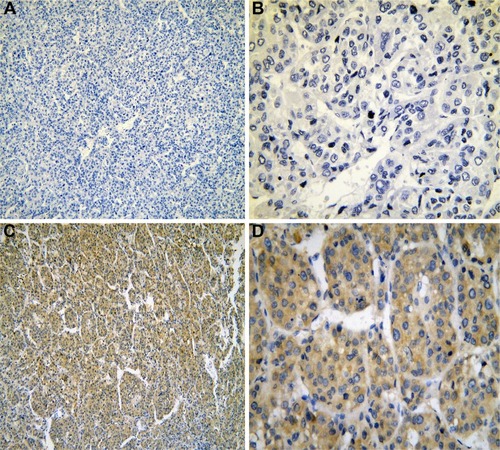
Figure 6 The expression of IRAK1 in IHC.
Abbreviations: IHC, immunohistochemistry; HCC, hepatocellular carcinoma.
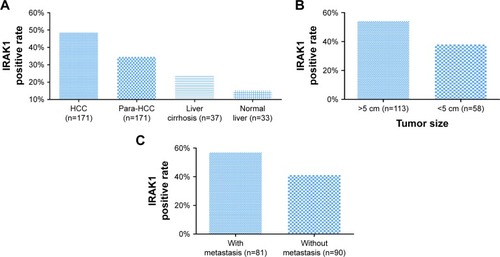
Table 1 Expression of IRAK1 in different types of liver tissues
Figure 7 The ROC curves of IRAK1 expression by IHC in HCC.
Abbreviations: ROC, receiver operating characteristic; IHC, immunohistochemistry; HCC, hepatocellular carcinoma; AUC, area under the curve.
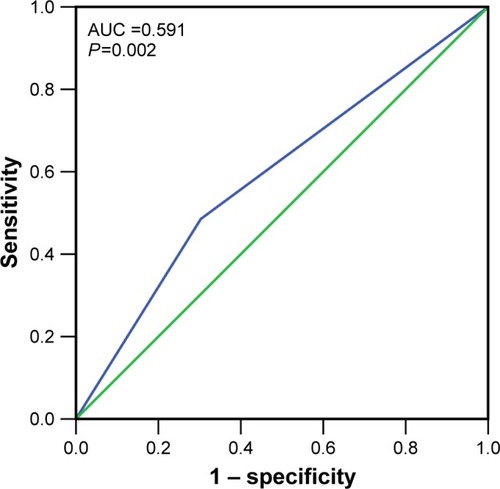
Table 2 Relationship between IRAK1 expression and clinicopathological features in HCC
Figure 8 The expression of IRAK1in HCC in the TCGA data.
Abbreviations: HCC, hepatocellular carcinoma; TCGA, the cancer genome atlas.
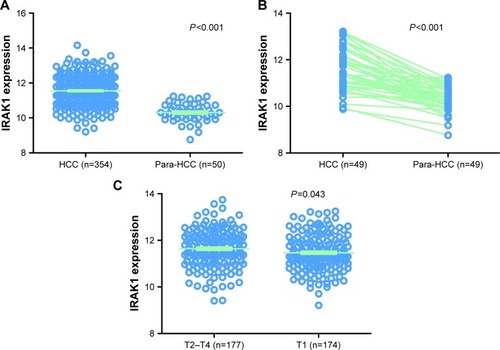
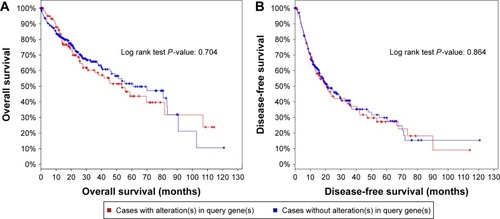
Figure 9 The ROC curve and Kaplan–Meier survival curve of IRAK1 expression for HCC from TCGA data.
Abbreviations: ROC, receiver operating characteristic; HCC, hepatocellular carcinoma; TCGA, the cancer genome atlas; AUC, area under the curve; Cum, cumulative.
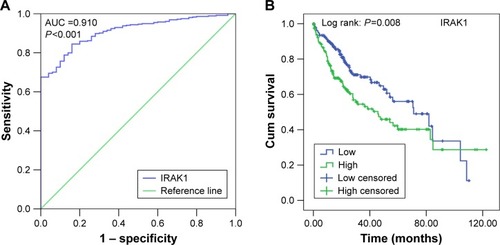
Table 3 Expression of IRAK1 detected by RNA sequencing and clinicopathological parameters in HCC in TCGA
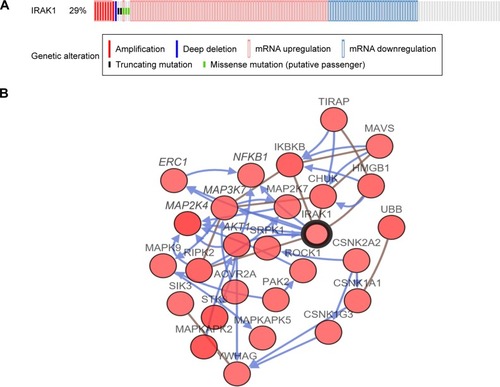
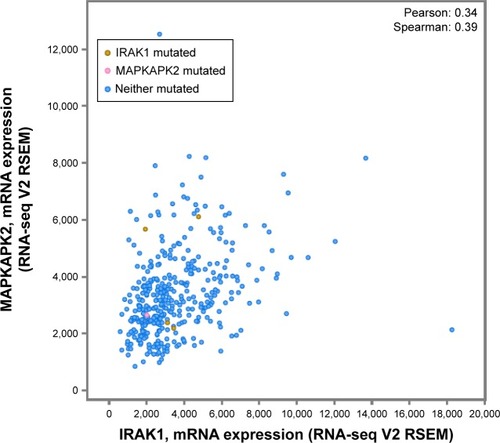
Figure 10 The alteration of IRAK1 and its interaction in altered neighboring genes in 440 HCC cases from the cBioPortal database.
Abbreviation: HCC, hepatocellular carcinoma.