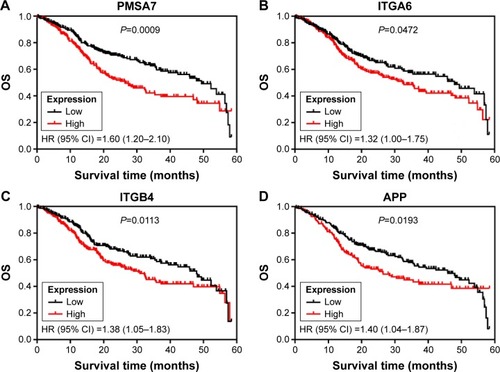
Figures & data
Table 1 Functional and pathway enrichment analysis of upregulated and downregulated DEGs in HNSCC
Figure 1 PPI network of differentially expressed genes.
Abbreviations: PPI, protein–protein interaction; DEGs, differentially expressed genes.
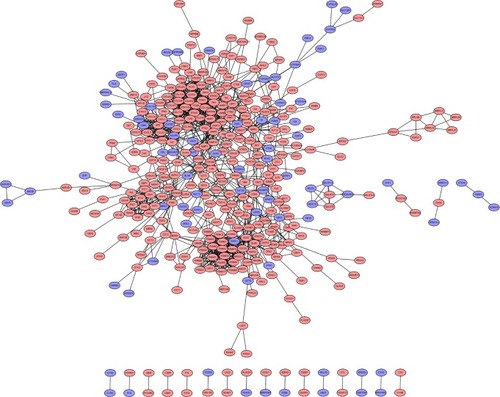
Figure 2 Functional modules in the PPI network.
Abbreviations: PPI, protein–protein interaction; DEGs, differentially expressed genes; MCODE, Molecular Complex Detection.
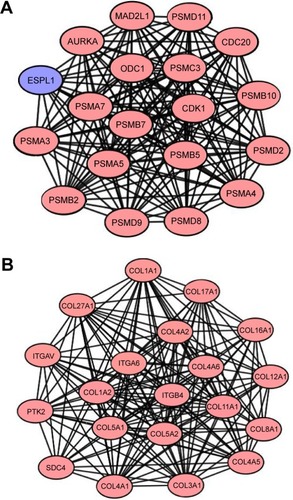
Table 2 The hub genes that had a degree >22 in PPI network
Table 3 Functional and pathway enrichment analysis of the DEGs in modules