Figures & data
Figure 2 Venn diagram of the DEGs in GSE46602 and GSE55945 (A) and fold change correlation between the two datasets (B).
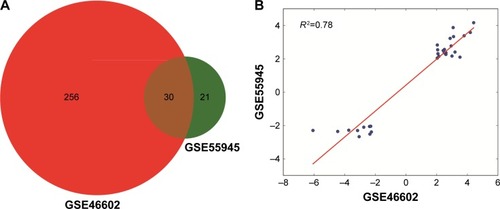
Figure 3 Heat map showing the expression pattern of 30 common DEGs in all tumor and normal samples.
Abbreviation: DEGs, differentially expressed genes.
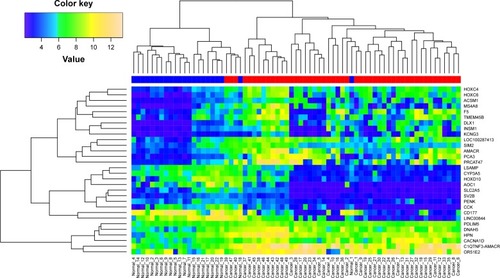
Table 1 The identified 30 common DEGs in GSE46602 and GSE55945
Table 2 KEGG pathway and GO enrichment results for the common DEGs
Figure 4 Regulation network between the identified common DEGs and reported miRNAs.
Abbreviation: DEGs, differentially expressed genes.
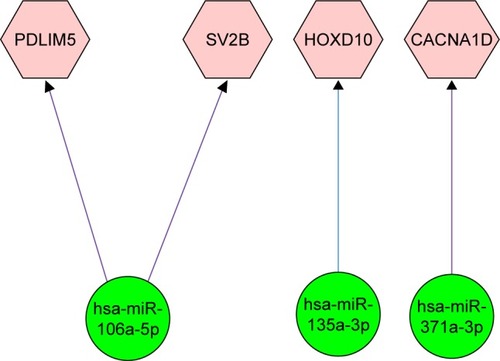
Figure 5 Kaplan–Meier curves of GSE16560 dataset (A) and GSE40272 (B).
Abbreviation: CI, concordance index.
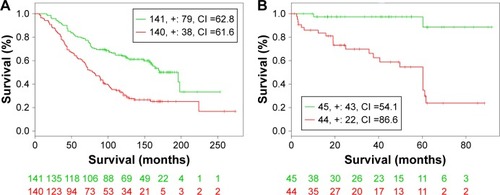
Table 3 Virtual validation results of prognostic performance