Figures & data
Figure 1 Profile of TPX2 expression in prostate cancer and human cancer cells.
Abbreviations: Breast ca., breast cancer; PRAD, prostate adenocarcinoma; Prostate ca., prostate cancer; RT-PCR, reverse transcription-polymerase chain reaction; TCGA, The Cancer Genome Atlas; H, TPX2 high expression; L, TPX2 low expression.
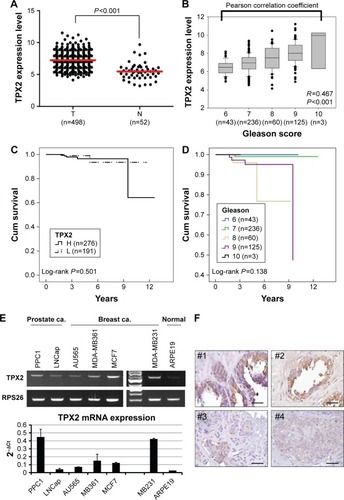
Figure 2 Inhibition of cell growth and reduction of tumorigenesis in human prostate cancer cell lines via TPX2 silencing.
Abbreviation: si-CTL, small interfering negative control.
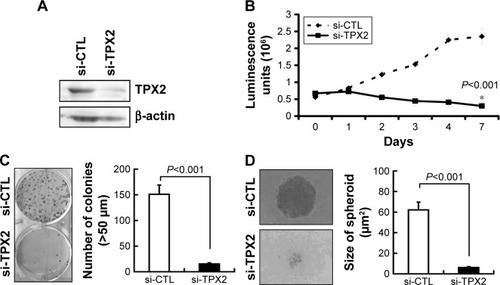
Figure 3 Increase of sub-G1 and reduction of G1 to S phase cells via TPX2 silencing.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; Noc, nocodazol; si-CTL, small interference-negative control; Thy, thymidine.
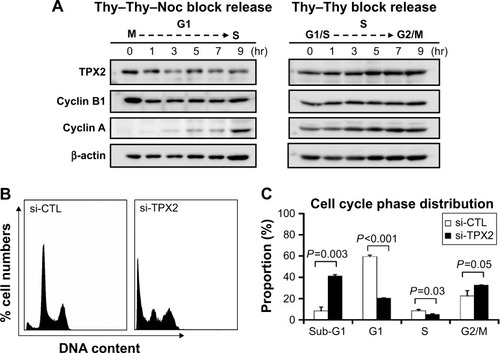
Figure 4 PPC1 cell apoptosis caused by TPX2 silencing.
Abbreviations: annexin V(+), annexin V positive; PI, propidium iodide; si-CTL, small interference-negative control; BMP, a image format of windows bitmap.
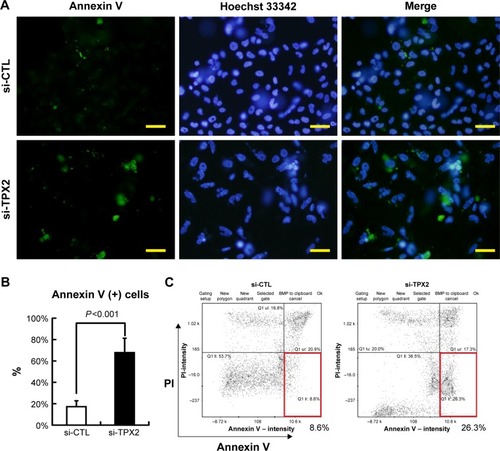
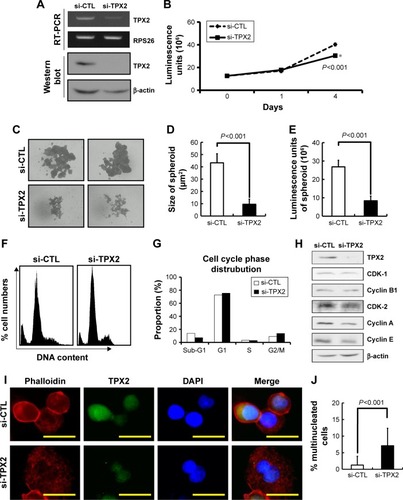
Figure 5 Depletion of TPX2, as reflected by changes in cell cycle regulation.
Abbreviations: qPCR, quantitative polymerase chain reaction; si-CTL, small interference-negative control.
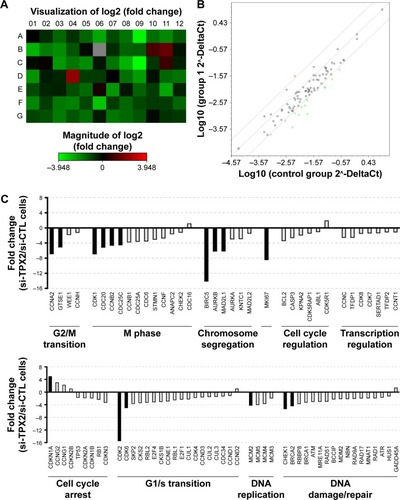
Figure 6 Increase in genomic instability via TPX2 depletion, resulting in micronucleation and DNA damage in PPC1 cells.
Abbreviations: CDK, cyclin-dependent kinase; DAPI, 4′,6-diamidino-2-phenylindole; si-CTL, small interference-negative control.
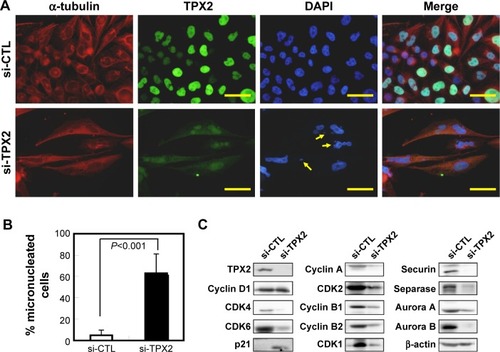
Figure 7 TPX2 depletion changes the cell fitness in LNCap prostate cancer cell line.
Abbreviations: CDK, cyclin-dependent kinase; DAPI, 4′,6-diamidino-2-phenylindole; si-CTL, small interference-negative control.