Figures & data
Table 1 Characteristics of 15 NSCLC patients with elevated level of KIF23
Figure 1 Relative expression of KIF23 in NSCLC and noncancerous lung samples from the study.
Abbreviations: TU, tumor; NC, non cancerous; NSCLC, non-small-cell lung cancer.
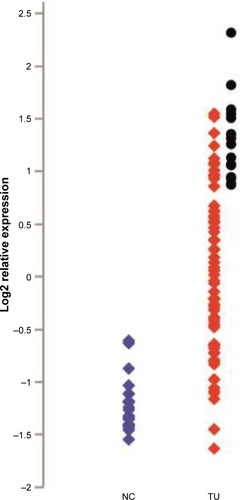
Table 2 KIF23 mutations in cancer
Figure 2 Sequences of case-specific heterozygous DNA variants present in 3 of 15 NSCLC samples.
Abbreviation: NSCLC, non-small-cell lung cancer.
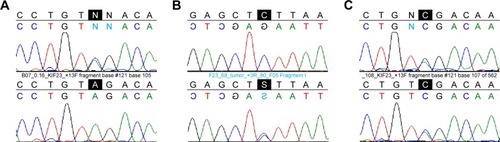
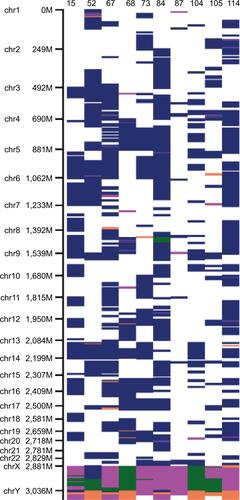
Figure 3 DNA CNA of chromosome 15 profiles in NSCLC. Whole-genome genotyping was performed using Illumina, SNP-array. Analysis was done using GenomeStudio and cnvPartition v. 3.2.0 with manual revision of the SNP-array profiles. BAFs for SNPs along the chromosome 15 show additional copy of chromosome 15 in 9 NSCLC cases. Only case 87 demonstrates normal profile with BAF =0.5 indicating absence of CNA on chromosome 15.

Figure S1 (A) Sequencing of cell cycle homology region (CHR) in the promoter of KIF23 in 9 NSCLC tumors. The consensus wild-type sequence TTTGAAA was present in all samples. (B) Correlation between TP53 and KIF23. Scatter plot of log2 mean-centered expression values of KIF23 and TP53, TP53: Pearson’s correlation 0.256, uncorrected p-value =0.008. Pearson product-moment correlation (r) and uncorrected p-value were calculated using R software package.
Abbreviation: NSCLC, non-small-cell lung cancer.
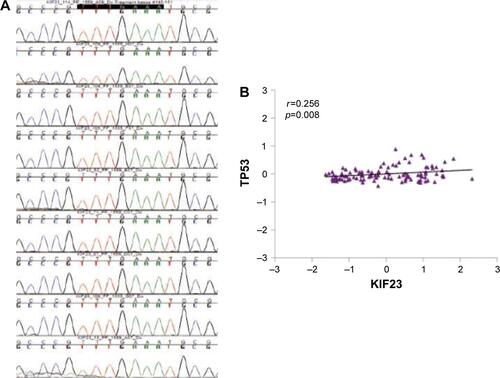
Table S1 Sanger sequencing and CNAs detection in NSCLC
Table S2 Chromosomal copy number alterations in NSCLC analyzed by genome wide genotyping using Illumina SNP-array