Figures & data
Figure 1 Differential expression of LGI3 and its receptors in glioma tissues.
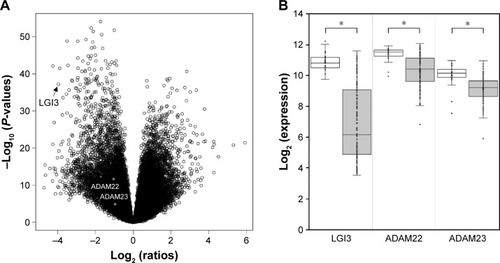
Figure 2 Comparative analysis of the glioma-altered gene products and LGI3-regulated genes.
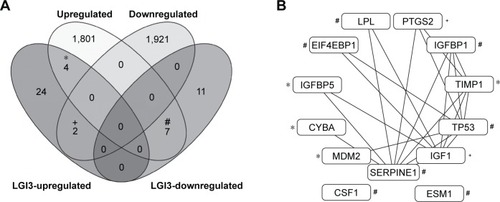
Table 1 Functional enrichment analysis of LGI3-regulated genes that are changed in glioma
Table 2 KEGG pathway analysis of LGI3-regulated genes that are changed in glioma
Figure 3 Associations between expression microarray analyses of LGI3 (A), ADAM22 (B) and ADAM23 (C) and glioma prognosis in patient cohorts.
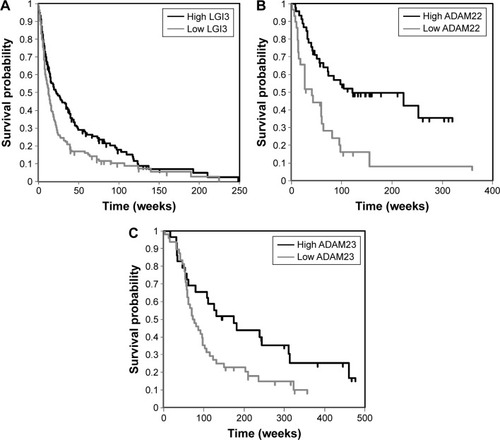
Table 3 Dataset summary of expression microarray analyses for glioma studies
