Figures & data
Figure 1 Schematic representation of enzymatic activity and linear structure of human LSD1 and LSD2.
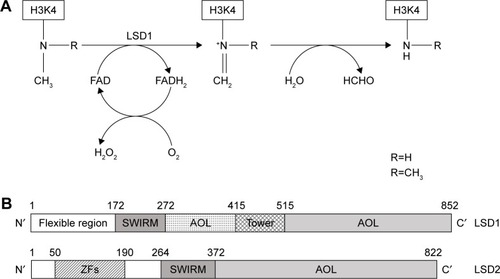
Table 1 Main LSD1 inhibitors introduced into a plan of clinical development
Figure 2 Schematic representation of the DOT1L protein.
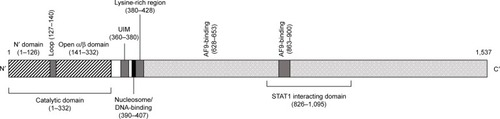
Table 2 Main properties of DOT1L inhibitors
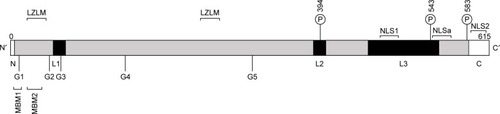
Figure 3 Linear structure of human menin.