Figures & data
Figure 1 Relative circRNA0003906 expression in CRC cell lines.
Abbreviations: CRC, colorectal cancer; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation.
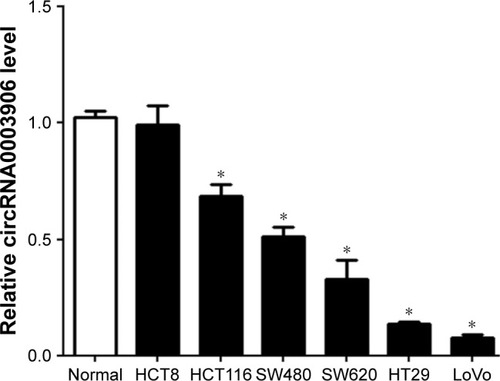
Figure 2 Relative circRNA0003906 expression in CRC tissues.
Abbreviations: CRC, colorectal cancer; SD, standard deviation.
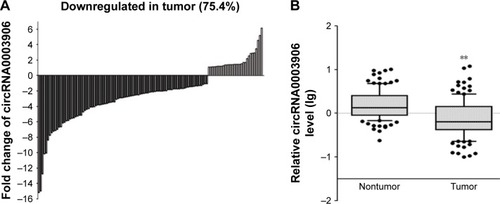
Table 1 Clinicopathological correlation of circRNA0003906 expression in CRC
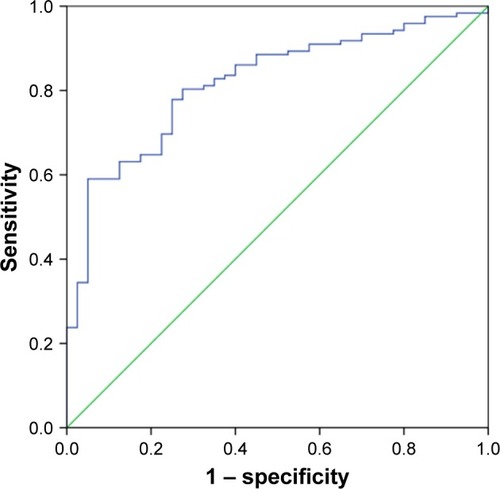
Figure 3 The ROC curve for differentiating CRC tissues from healthy controls.
Abbreviations: CRC, colorectal cancer; ROC, receiver operating characteristics.