Figures & data
Table 1 The top 10 dysregulated mRNAs, miRNAs, lncRNAs
Figure 1 Hierarchical clustering dendrograms of identified co-expressed genes in modules.
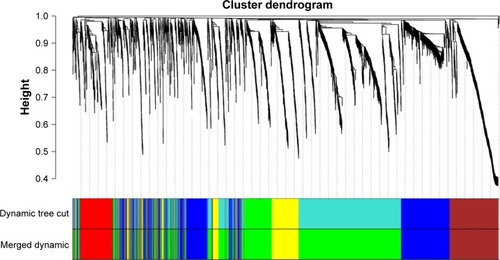
Figure 2 PPI network visualization of modules identifies hub genes.
Abbreviations: MMP, matrix metalloproteinase; PPI, protein–protein interaction.
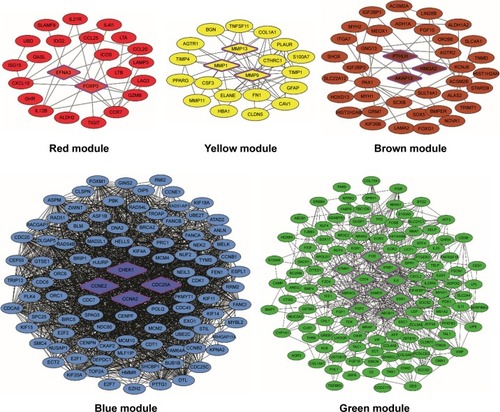
Figure 3 Module–trait relationships.
Abbreviation: ME, module eigengenes.
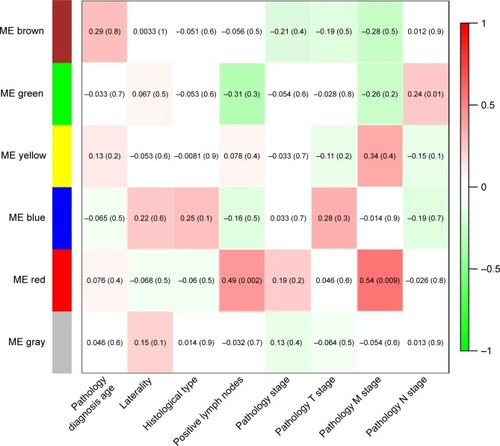
Table 2 The clinical features of triple negative breast cancer patients
Figure 4 Network diagram.
Abbreviations: ceRNA, competing endogenous RNA; lncRNA, long non-coding RNA; miRNA, microRNA; TNBC, triple negative breast cancer.
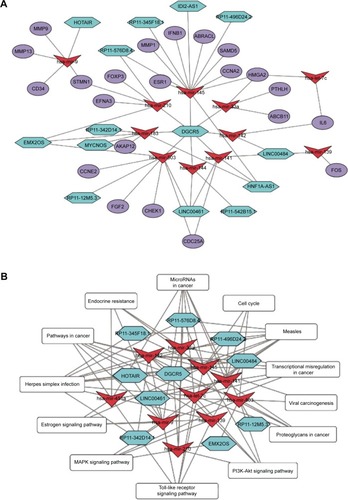
Table 3 Each node of ceRNA crosstalk network
Table 4 KEGG annotation of ceRNA crosstalk network
Figure 5 Survival curve analysis of ceRNA network for overall survival in triple negative breast cancer.
Abbreviations: ceRNA, competing endogenous RNA; DEGs, differentially expressed genes; miRNA, micro RNA.
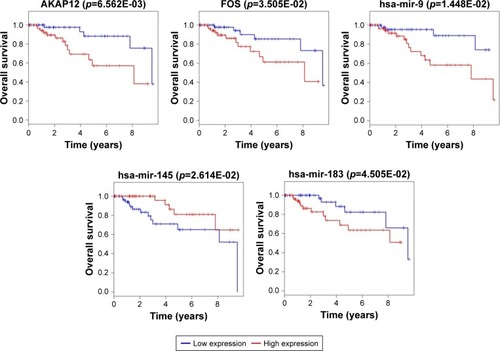
Figure 6 Survival curve analysis of ceRNA network for the overall survival in triple negative breast cancers.
Abbreviations: ceRNA, competing endogenous RNA; lncRNA, long non-coding RNA.
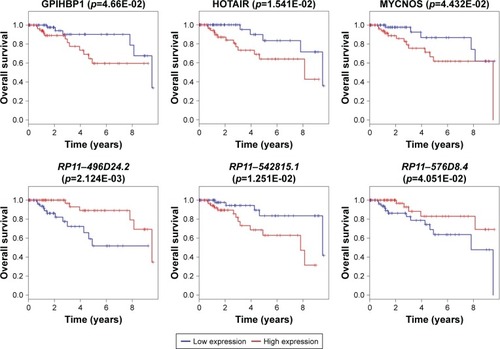
Table S1 The enriched KEGG pathways for each module
