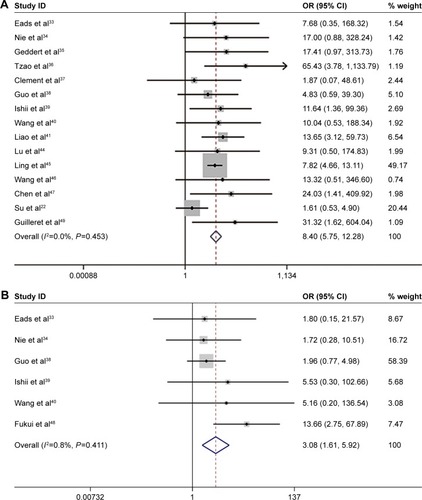
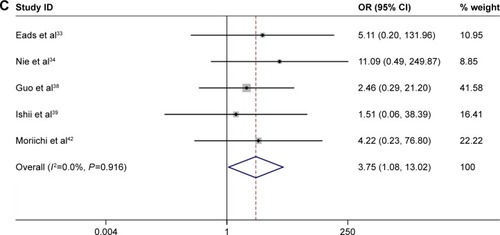
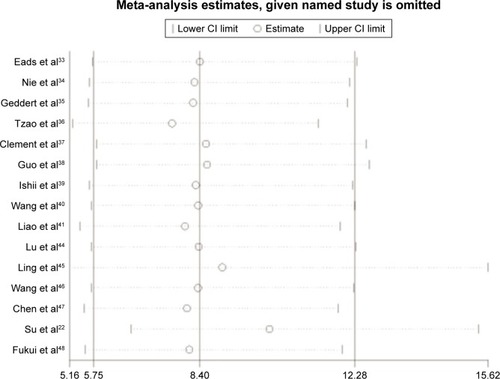
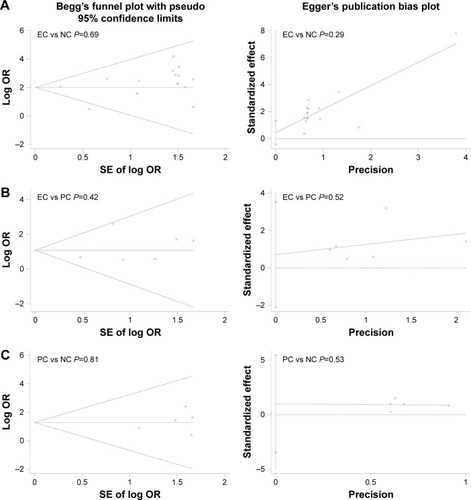
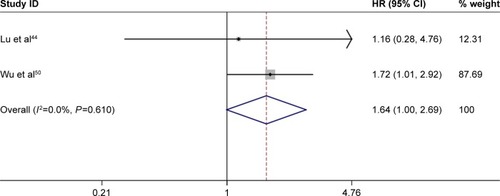
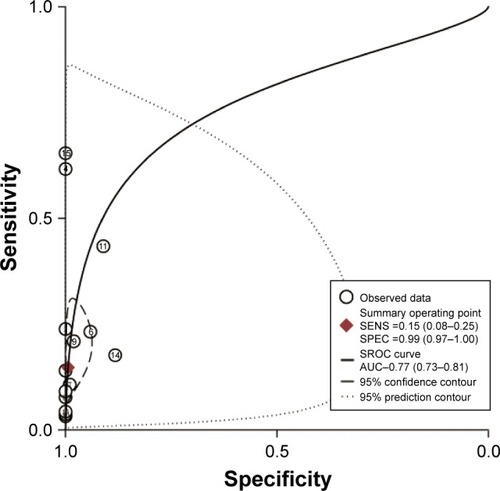
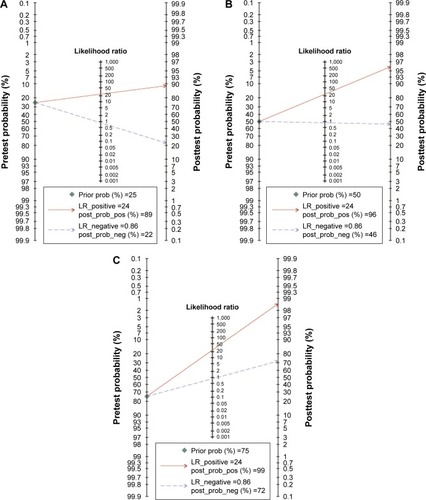
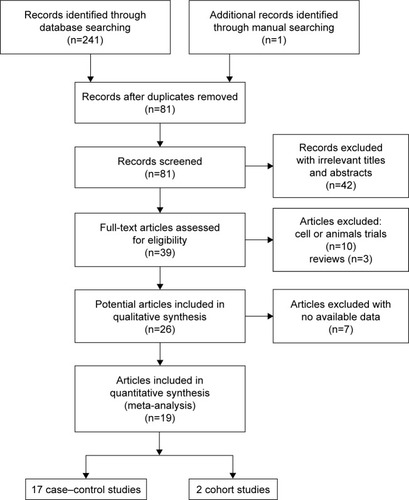
Figures & data
Table 1 The basic characteristics of all eligible studies
Table 2 Subgroup analyses of MLH1 promoter methylation in esophageal cancer
Table 3 Association between MLH1 promoter methylation and clinicopathological features of esophageal cancer patients