Figures & data
Table 1 Summary of baseline patient characteristics
Figure 1 Baseline somatic gene aberrances identified by NGS.
Abbreviations: chemo, chemotherapy; CN_amp, copy number amplification; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; Mut, mutation; NA, not available; NGS, next-generation sequencing; TKI, tyrosine kinase inhibitor.
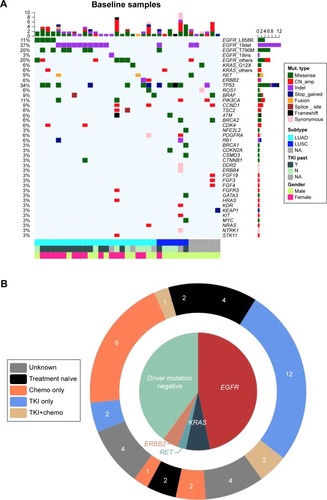
Figure 2 IGV screenshots displaying the reads of each mutation subtype identified by NGS.
Abbreviations: IGV, Integrative Genomics Viewer; NGS, next-generation sequencing.
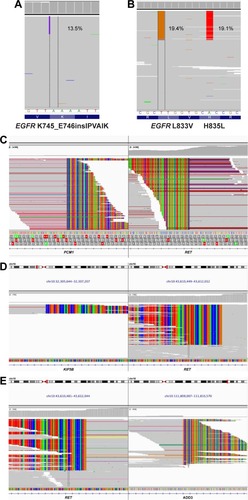
Figure 3 Correlation of patient baseline characteristics, baseline somatic aberrances, and PFS.
Abbreviations: AF, allelic fraction; MTT, matched targeted therapy; PFS, progression-free survival.
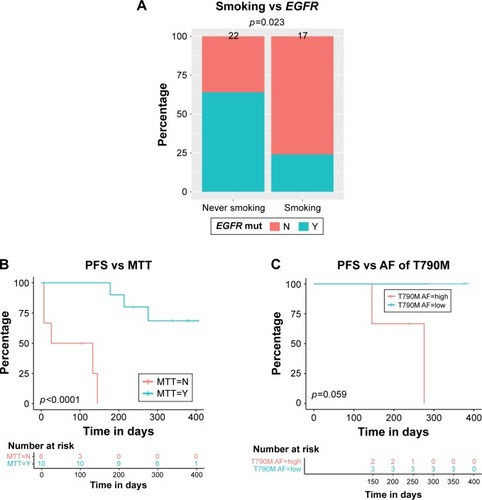
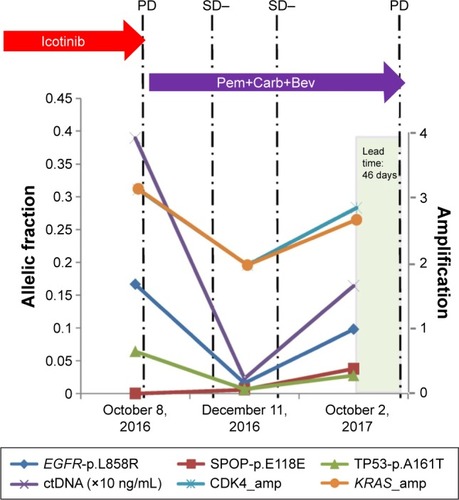
Figure 4 A lung squamous cell carcinoma patient is presented here to illustrate the value of ctDNA in predicting disease progression earlier than radiographic imaging.
Abbreviations: amp, amplification; ctDNA, circulating tumor DNA; PD, progressive disease; Pem+Carb+Bev, pemetrexed+carboplatin+bevacizumab; SD, stable disease.