Figures & data
Table 1 Clinical characteristics of patients
Table 2 Correlation between GAPDH level (raw Ct value) in human pleural effusion and clinicopathological factors of patients with BPE and LC-MPE
Figure 1 The stability of GAPDH in human pleural effusion.
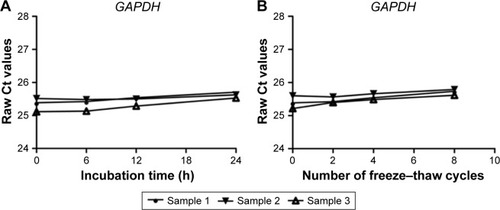
Figure 2 Comparison of pleural fluid levels of MALAT1 (A), H19 (B), CUDR (C), and PANDAR (D) in LC-MPE and BPE.
Abbreviations: LC-MPE, lung cancer-associated malignant pleural effusion; BPE, benign pleural effusion; lncRNAs, long noncoding RNAs.
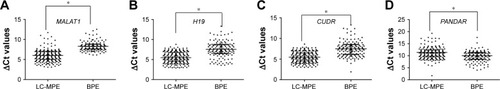
Figure 3 The stability of lncRNAs in human pleural effusion.
Abbreviation: lncRNAs, long noncoding RNAs.
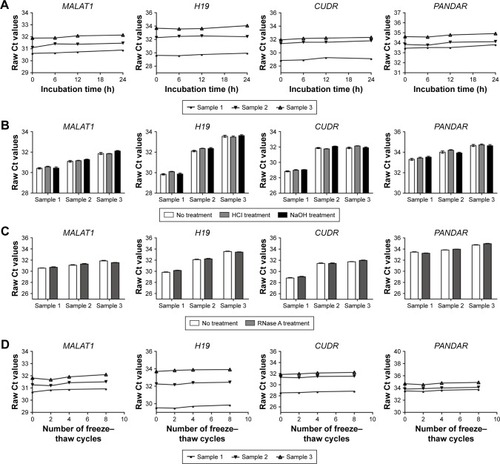
Table 3 Performance of pleural effusion lncRNAs and CEA in the differential diagnosis of LC-MPE from BPE
Figure 4 Evaluation of pleural fluid lncRNAs for the diagnosis of LC-MPE.
Abbreviations: LC-MPE, lung cancer-associated malignant pleural effusion; BPE, benign pleural effusion; lncRNAs, long noncoding RNAs; AUCs, area under the curves; CEA, carcinoembryonic antigen.
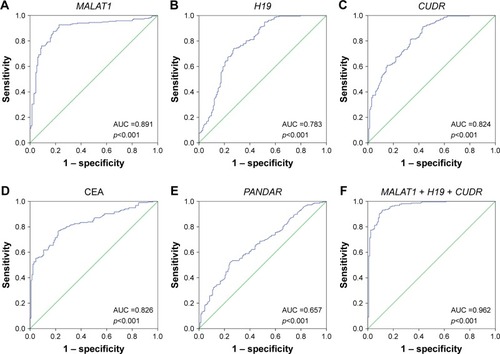
Figure 5 ROC curves to compare the ability of CEA, MALAT1, and a combination of CEA and MALAT1, to discriminate LC-MPE from BPE.
Abbreviations: LC-MPE, lung cancer-associated malignant pleural effusion; BPE, benign pleural effusion; AUC, area under the curve; ROC, receiver operating characteristic; CEA, carcinoembryonic antigen.
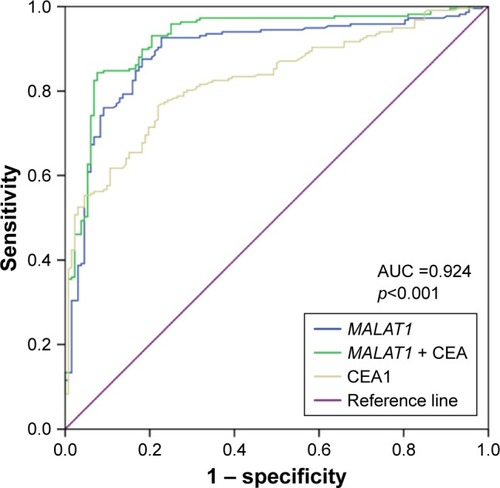
Table S1 Primers sequences list
