Figures & data
Figure 1 Molecular mechanisms of cisplatin resistance.
Notes: Multiple cellular alterations in cancer cells, including cell cycle, apoptosis, autophagy, stemness, intracellular detoxification, and drug influx/efflux, contribute to cisplatin chemoresistance through genetic and/or epigenetic regulation of multiple signaling pathways. Some major genetic and epigenetic factors are illustrated in the figure (see text for detailed discussion).
Abbreviations: ALDH1, aldehyde dehydrogenase 1 family member A1; ATG7, autophagy associated gene; BRCA2, breast cancer susceptibility proteins 2; CTR1, copper transporter 1; ERCC1, excision repair cross-complementing rodent repair deficiency, complementation group 1; GSH, glutathione; GST, glutathione S-transferase; HR, homologous recombination; MMR, mismatch repair; MRP, multidrug-resistant-associated protein; NER, nucleotide excision repair; γ-GCS, γ-glutamylcysteine synthetase.
Abbreviations: ALDH1, aldehyde dehydrogenase 1 family member A1; ATG7, autophagy associated gene; BRCA2, breast cancer susceptibility proteins 2; CTR1, copper transporter 1; ERCC1, excision repair cross-complementing rodent repair deficiency, complementation group 1; GSH, glutathione; GST, glutathione S-transferase; HR, homologous recombination; MMR, mismatch repair; MRP, multidrug-resistant-associated protein; NER, nucleotide excision repair; γ-GCS, γ-glutamylcysteine synthetase.
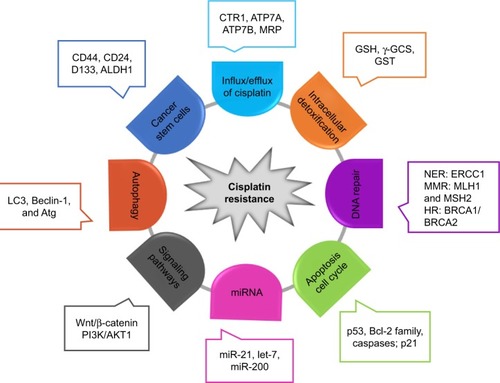
Table 1 Predictive lncRNAs involved in response to cisplatin
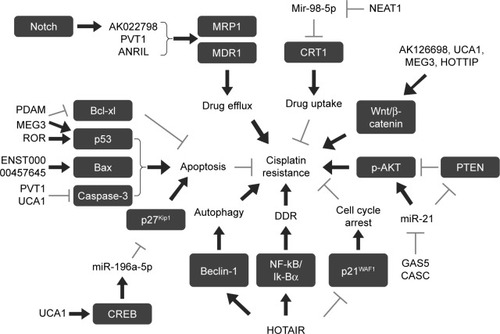
Figure 2 Role of lncRNAs in cisplatin resistance.
Notes: lncRNAs that regulate drug efflux, drug uptake, apoptosis, DDR, cell cycle arrest, and autophagy of cancer cells are implicated in cisplatin resistance. Gray arrows indicate inhibition and black arrows indicate activation.
Abbreviations: DDR, DNA damage response; lncRNAs, long non-coding RNAs; MDR1, multidrug-resistant protein; MRP1, multidrug-resistant-associated protein 1.
Abbreviations: DDR, DNA damage response; lncRNAs, long non-coding RNAs; MDR1, multidrug-resistant protein; MRP1, multidrug-resistant-associated protein 1.