Figures & data
Figure 1 Aerobic glycolysis is involved in regulating hepatoma cell proliferation by miR-342-3p.
Abbreviation: NC, negative control.
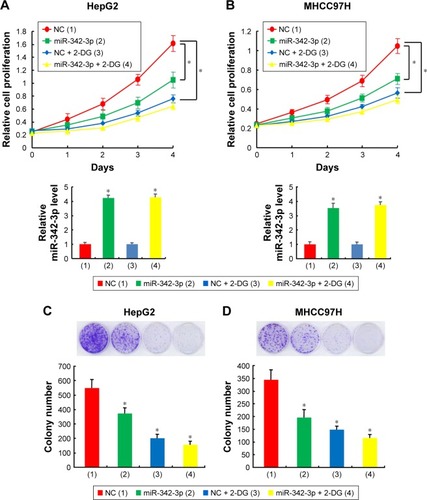
Figure 2 IGF-1R is a direct target of miR-342-3p.
Abbreviations: NC, negative control; 3′-UTR, 3′-untranslated region.
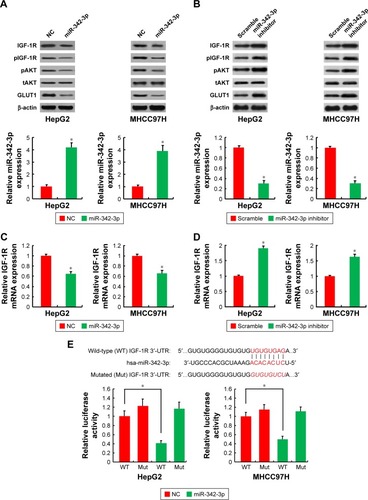
Figure 3 miR-342-3p suppresses hepatoma cell proliferation and glycolysis mainly through inhibition of IGF-1R expression.
Abbreviations: NC, negative control; CCK-8, cell counting kit-8; ECAR, extracellular acidification rate; OCR, oxygen consumption rate; FCCP, p-trifluoromethoxy carbonyl cyanide phenylhydrrazone; Rote/AA, rotenone plus the mitochondrial complex III inhibitor antimycin A.
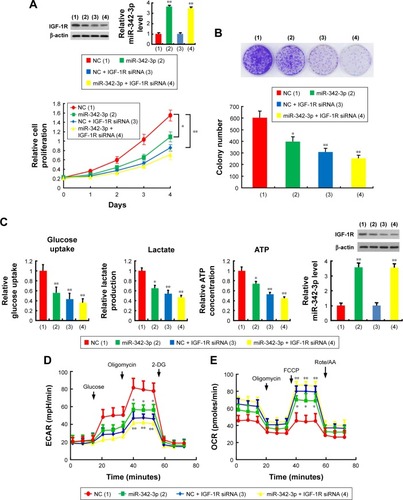
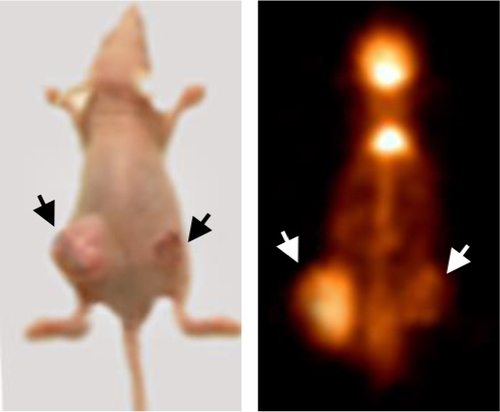
Figure 4 miR-342-3p/IGF1R axis regulates glycolysis and tumor growth in vivo.
Abbreviations: FDG microPET, fluorodeoxyglucose micropositron emission tomography; T/M, tumor to muscle; NC, negative control.
Figure S1 Screening for the target of miR-342-3 in HEK293T cells.
Abbreviation: NC, negative control.
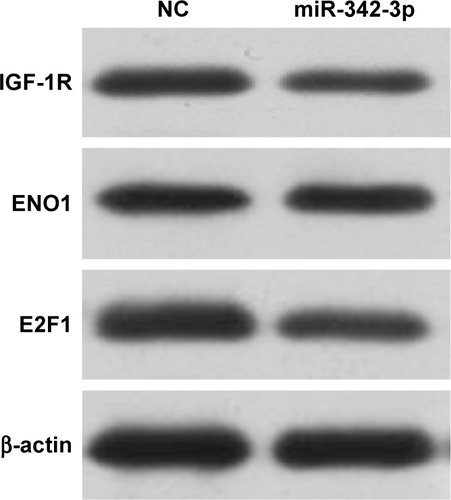
Figure S2 miR-342-3p suppresses proliferation, glucose uptake and the production of lactate and ATP through inhibition of IGF-1R expression in HCC cells.
Abbreviations: NC, negative control; CCK-8, cell counting kit-8.
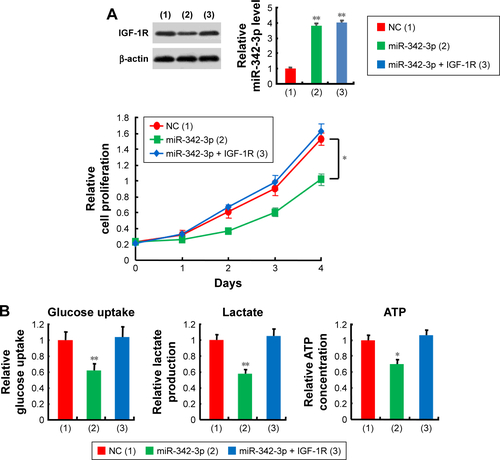
Figure S3 miR-342-3p/IGF-1R axis regulates glycolysis and tumor growth in vivo.
Abbreviation: FDG microPET, fluorodeoxyglucose micropositron emission tomography.