Figures & data
Table 1 Characteristics of the microarray datasets from GEO database
Figure 1 Venn diagram of DEM/DEG selection in different datasets.
Abbreviations: DEGs, differentially expressed genes; DEM, differentially expressed miRNA; miRNAs, microRNAs.
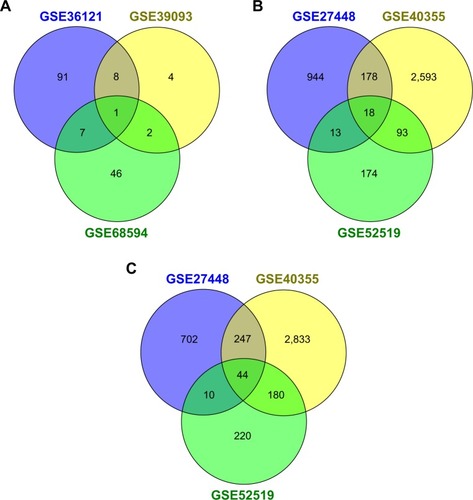
Table 2 Results of significant GO biological processes and the KEGG pathway analysis
Figure 2 PPI network of DEGs.
Abbreviations: DEGs, differentially expressed genes; PPI, protein–protein interaction.
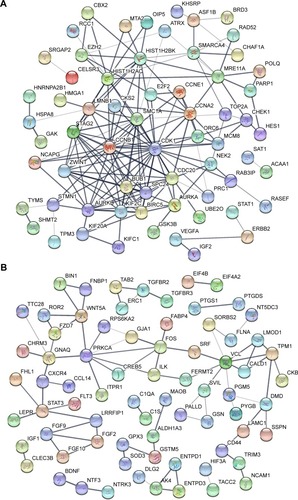
Table 3 The top 5% hub genes in the PPI networks
Figure 3 The miRNA–mRNA regulatory network.
Abbreviation: miRNA, microRNA.
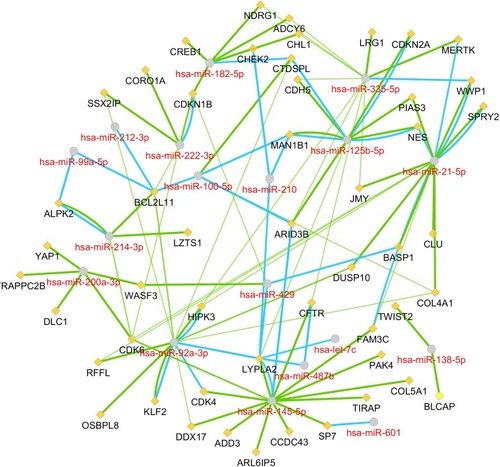
Table 4 Results of the significant DEM expression validation and the relationship with bladder cancer clinical stages in TCGA dataset
Figure 4 The Kaplan–Meier plots of patients from TCGA dataset.
Abbreviation: TCGA, The Cancer Genome Atlas.
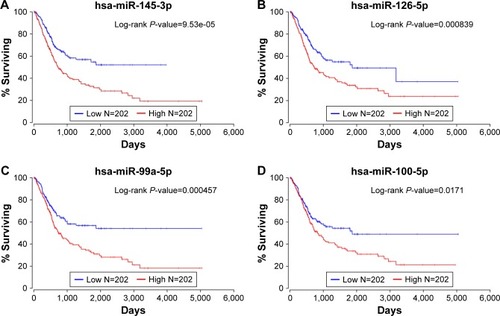
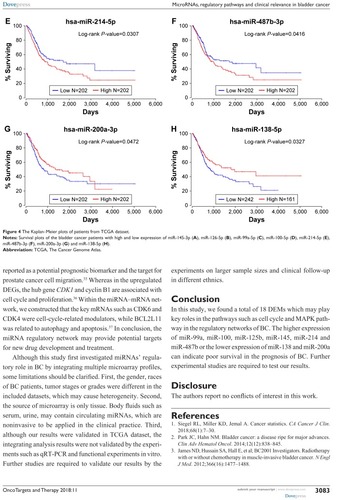
Figure S1 Venn diagram of DEMs (A: Ta–T1 vs T2–T4; B: G3 vs G1) in different datasets.
Abbreviations: DEMs, differentially expressed miRNAs; miRNAs, microRNAs.
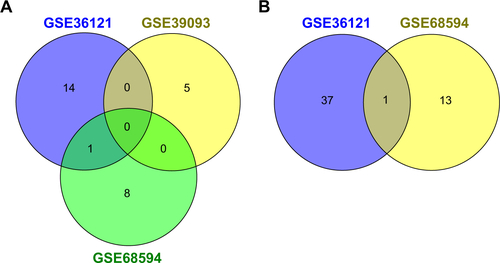
Table S1 Detailed information of patients in each included GEO datasets
